Dynamics of Structures and
Interactions of Macromolecules in Biology
We are part of the Red Blood Cell integrated biology lab
located at Hôpital Necker - Enfants Malades.
ABOUT US
DSIMB is a bioinformatics team in the Inserm unit UMR-S1134 part of the Laboratory of Excellence GR-Ex, whose focus is the study of membrane proteins involved in red blood cell. The long-term scientific goal of DSIMB is to elucidate the mechanisms governing important functions of proteins of the RBC membrane, using information from protein structure and dynamics. Both membrane protein structure and dynamics determine RBC function, but they are difficult to characterize experimentally. Our expertise in structural bioinformatics and molecular simulations enables us to tackle both aspects: the prediction of 3D protein structures and the exploration of their dynamics.
Part of our activity is devoted to the development of methodologies to characterize membrane systems. These methodologies aim at predicting structural and functional properties of membrane proteins from sequence and at elucidating dynamics and thermodynamics properties starting from 3D structures. We also apply those methodologies to specific cases of interest for our unit and for GR-Ex. We consider with particular interest systems relevant for diseases studied by GR-Ex members, and we plan to contribute to the design of drugs able to interfere with the disease mechanisms.
Our lab also works on the development of new methods for the prediction of protein 3D structures based on their amino acid sequence. New methods based on machine and deep learning techniques were and are currently being developed.
OUR TEAM CONTRIBUTES TO
- The development of bioinformatics tools for predicting structural and dynamics properties of proteins. Tools are available here.
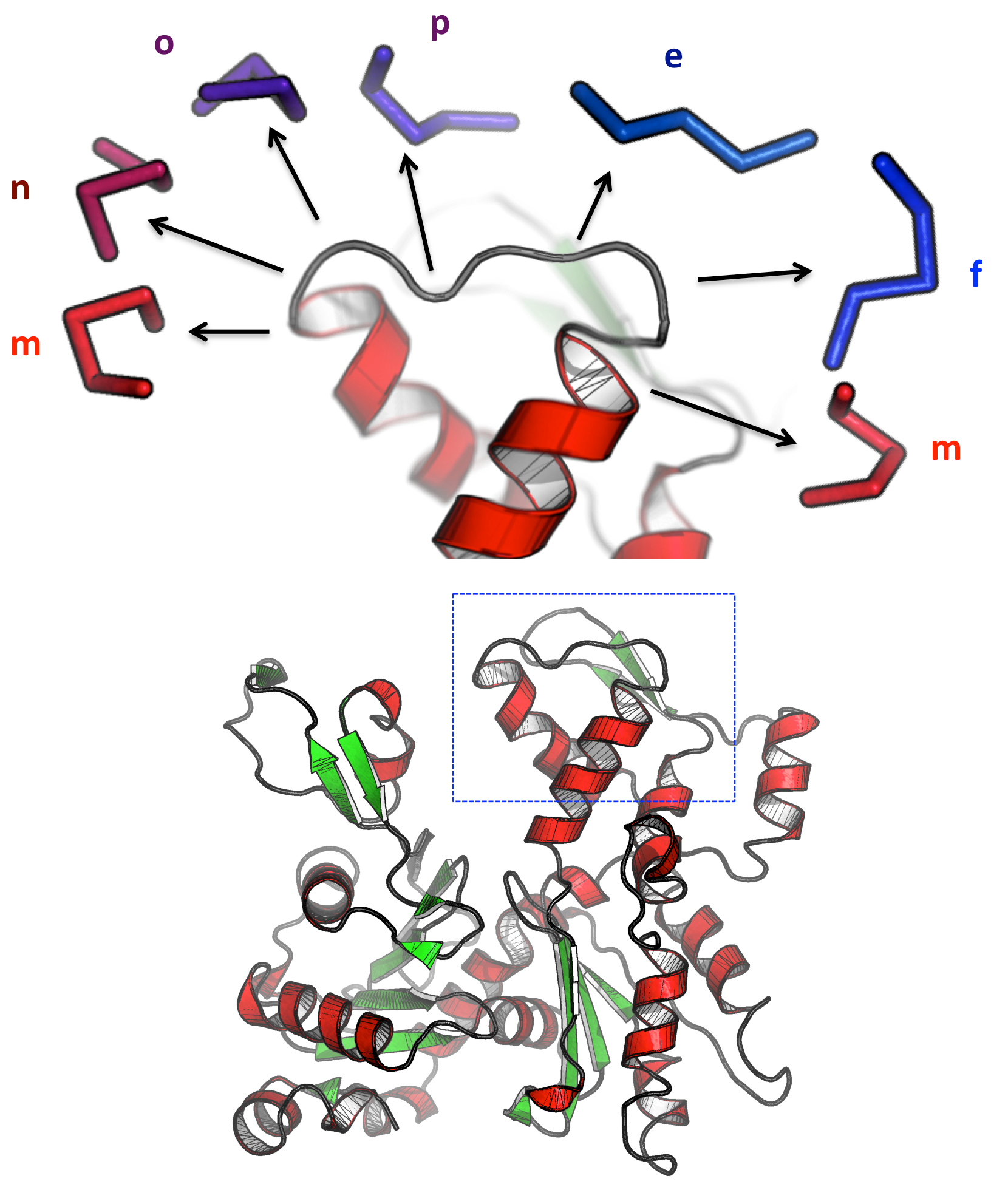
- A better understanding of Duffy Antigen/Receptor for Chemokines (DARC): These chemokine receptors are erythrocyte transmembrane receptors for Plasmodium knowlesi and Plasmodium vivax merozoites, two major agents of malaria. We elaborated 3D structural models for DARC using experimental data provided by partner 2. These models were further used to predict potential binding zones between DARC and CXCL8 natural ligand and P. vivax partners. Once identified, these zones could be targeted with new designed molecules preventing interaction.
- DARC VHHs: We modelled the structure of VHH sequences able to bind Nterminus of DARC and we identified important amino acids involved in VHH binding. This work was conducted in collaboration with O. Bertrand (Team 1) and Dr. Mr. Czerwinski, Wroclaw, Poland, on camel VHHs. This model system allows the development of new tools in transfusion medicine (agglutination tests in routine) and could serve as blocking agents against P. vivax and HIV.
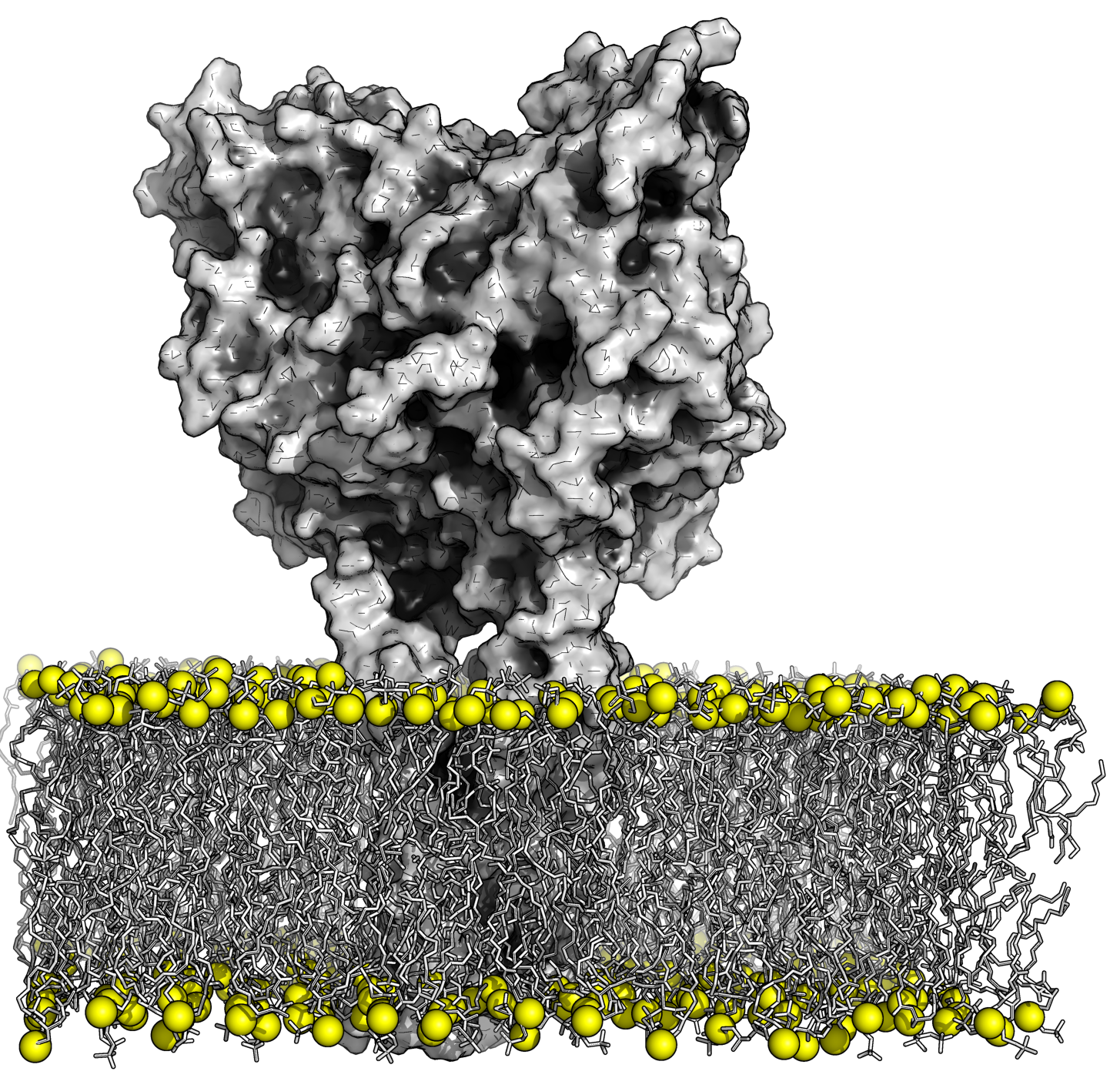
We will study transporter proteins in red blood cells with the aim at providing an insight into substrates transport mechanisms, understanding inhibition and predicting impact of mutations involved in pathological diseases, at an atomistic level. We will explore adhesion proteins in RBC in particular, integrins for gaining a molecular view on pathological cell adhesion to design anti-adhesive molecules.
EUROPEAN REGIONAL DEVELOPMENT FUND
The European Regional Development Fund (ERDF, FEDER in french) operates as part of the Economic, Social and Territorial Cohesion Policy. It aims to strengthen economic and social cohesion in the European Union by correcting imbalances between its regions. In France, the ERDF allocation is of 9.5 billion euros for the 2014-2020 programming period.


