Example
To use iPBAVizu follow the following steps or watch the VIDEO:
- 1 - BEFORE TO START
For the example below, we used the PDB structures 1JKW and 1VIN.
Launch PyMol program, and load the 2 structures 1JKW and 1VIN. You can use the "fetch" command in the PyMol console:fetch 1JKW 1VIN
- 2 - LAUNCH iPBAVizu
As soon as 2 structures or 2 chains are loaded and available in a current PyMol session, iPBAVizu can be launched from the PyMol Wizard menu.
iPBAVizu menu appears in PyMol the Measurement or Fit native functions.
Users can select 2 chains among the available loaded structures.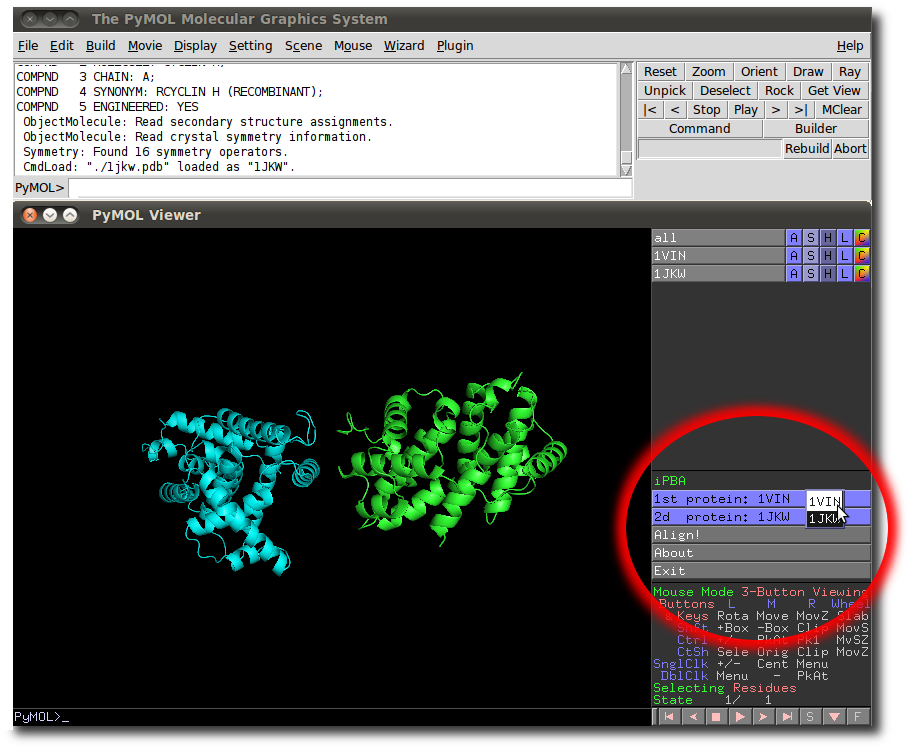
- 3 - RUN SUPERIMPOSITION
Next, by clicking on "Align!", iPBAVizu runs iPBA programs.
After a quick time (2-3 seconds depending on protein sizes), results are spawn in 2 new protein objects in PyMol and in a new independent window.
The 2 new objects are the 2 aligned structured.
The new independent window contains both scores (such like GDT_TS, RMSD ...) and an interactive sequence alignment manager. This latter displays both residue and protein block sequence from both aligned structures.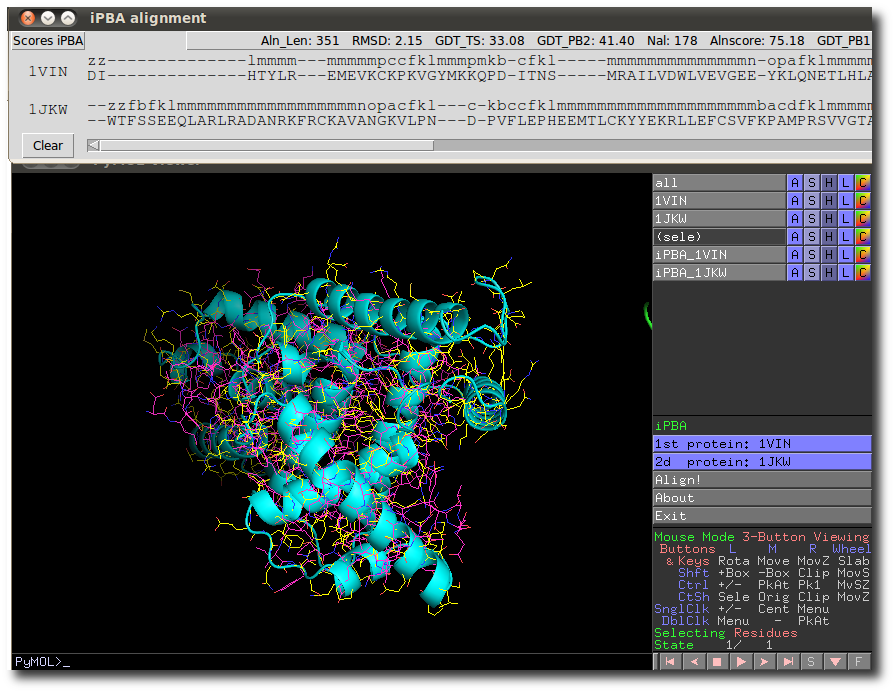
- 4 - EXPLORE SEQUENCE AND STRUCTURAL ALIGNMENT
Users can highlight in this manager any residue or protein block of one or both sequences.
Highlighting selects directly residue among the 2 new aligned protein objects created in the 3D PyMol window.
This interactive function provides a way to explore sequence and structural alignment.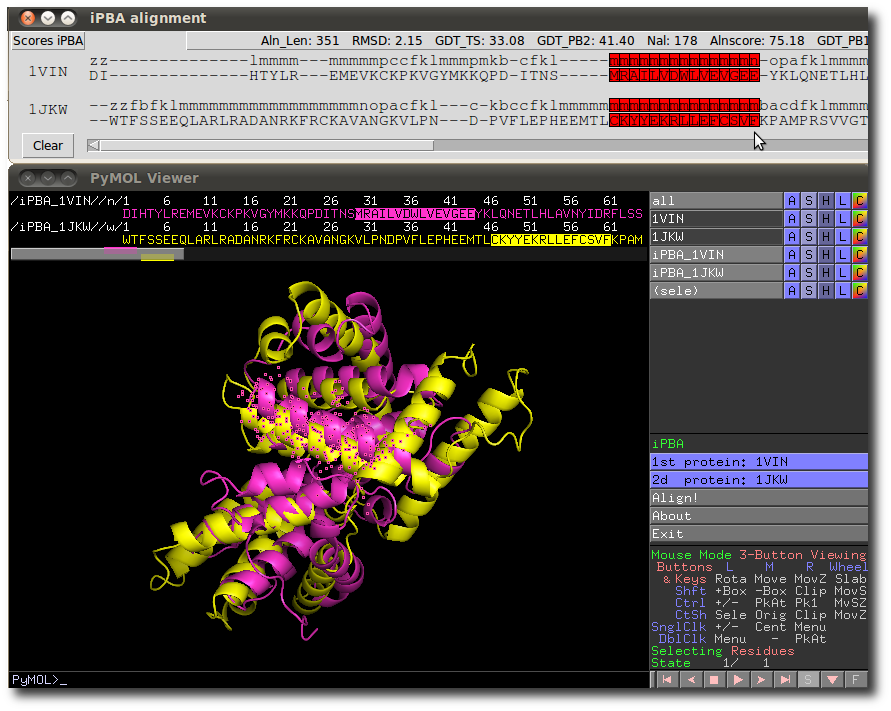
For more details, you can watch the following video.

