 |
MitoGenesisDB: Mitochondrial Spatio-Temporal Expression Database |
Biological Background Definitions References |
 |
MitoGenesisDB: Mitochondrial Spatio-Temporal Expression Database |
Biological Background Definitions References |
Mitochondria constitute complex and flexible cellular entities, which play crucial roles in normal and pathological cell conditions. The database MitoGenesisDB focuses on the dynamic of mitochondrial protein formation through global mRNA analyses. Three main parameters confer a global view of mitochondrial biogenesis:
| Available Tools | More Information ? |
|
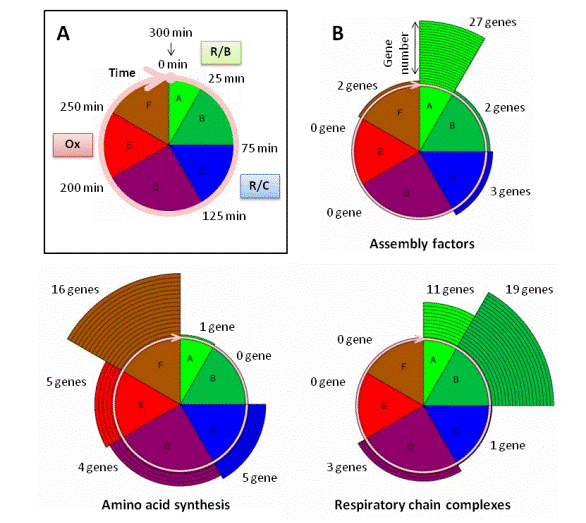
In MitoGenesisDB, we provide several graphical representations. One of them illustrate the concept of mitochondrial cycle (see A, figure on the left). The pie chart shows the correspondence between the different EDPM clusters (or phases A to F, see the Biological Background Section and Lelandais et al. (2009) for more information) and the major R/B, R/C and Ox phases identified in the 5-hour (or 300-minutes) Yeast Metabolic Cycle (YMC) (see the Biological Background Section and Tu et al. (2005) for more information). Each EDPM cluster comprises distinct subclasses of mitochondrial genes whose mRNAs peak in different time window of the YMC: between 0 and 25 minutes for genes in cluster A; between 25 and 75 minutes for genes in cluster B; between 75 and 125 minutes for genes in cluster C; between 125 and 200 minutes for genes in cluster D; between 200 and 250 minutes for genes in cluster E and finally between 250 and 300 minutes for genes in cluster F. As an illustration, repartitions among the mitochondrial cycle for genes that belong to the functional categories Assembly Factors, Respiratory Chain Complexes and Amino Acid Synthesis are presented in B (figure on the left).The number of genes in each class is shown with surrounding circular segments. This graphical representation allows to easily observing that the temporal groups A to F correlate with functional properties of the corresponding proteins. The first mRNAs to appear are those for proteins whose function is associated with translation machinery (or regulation) and assembly factors, followed by those involved in the synthesis of respiratory chain structural proteins and finally mRNAs coding enzymes involved in the amino-acid biosynthesis. |
 |