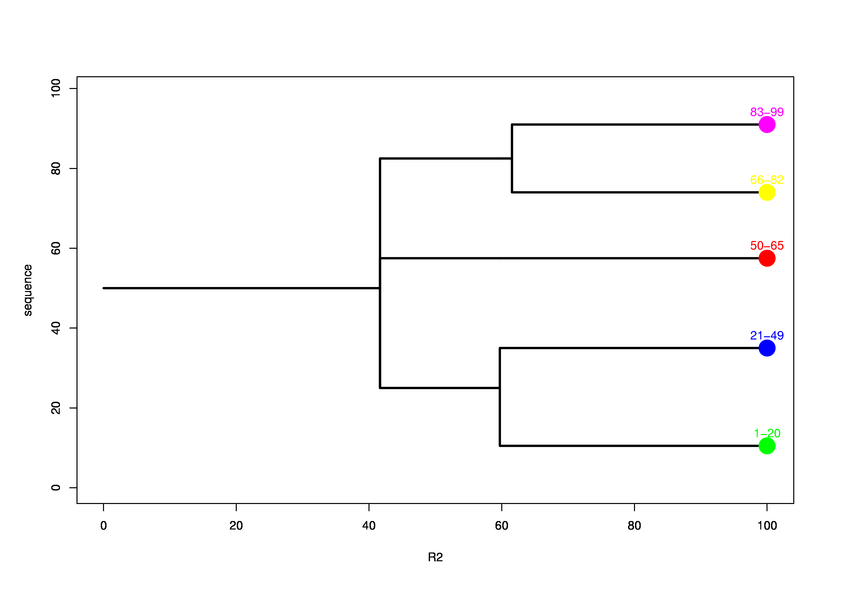
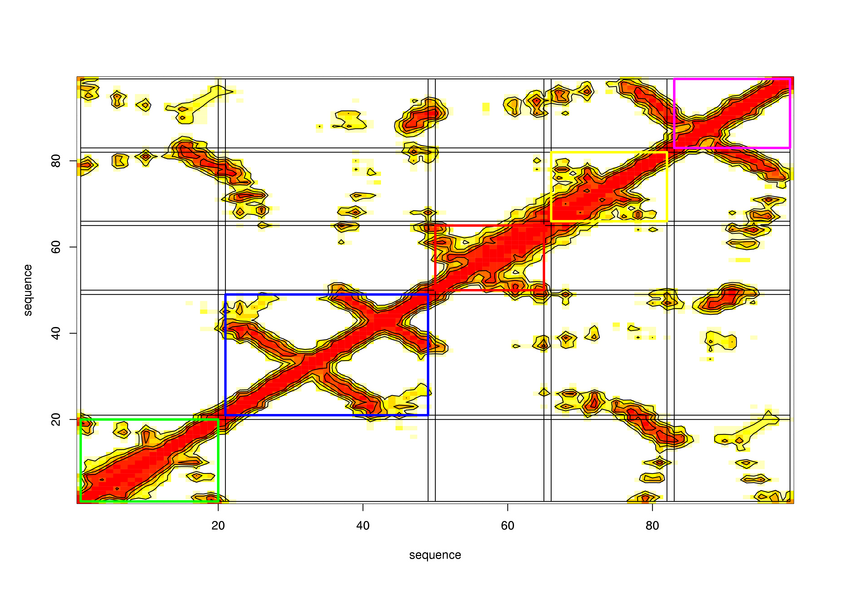
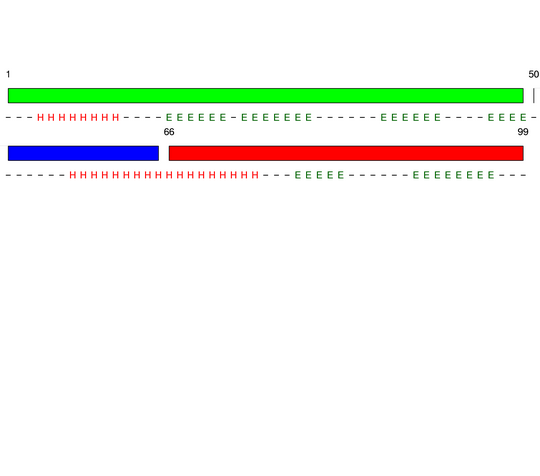
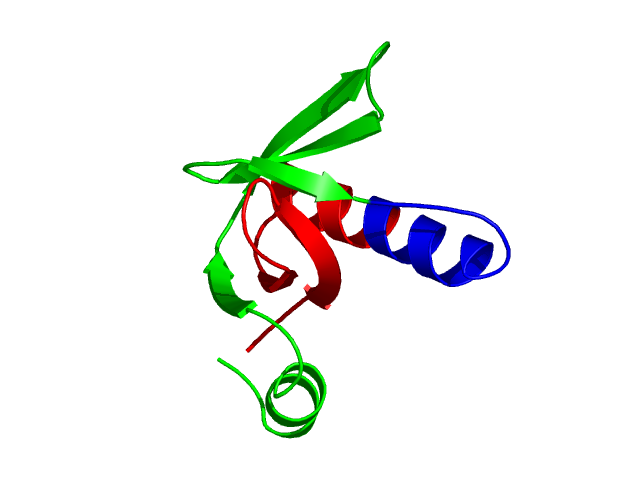
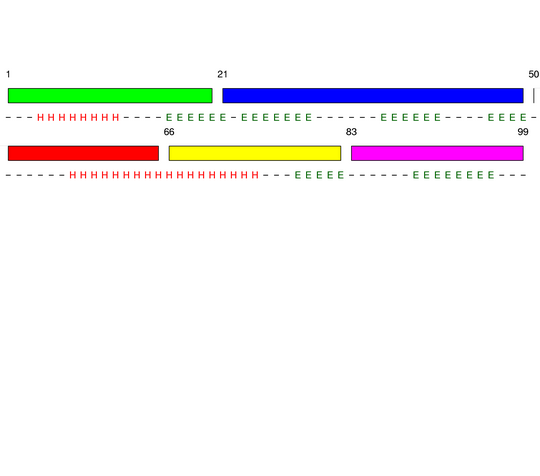
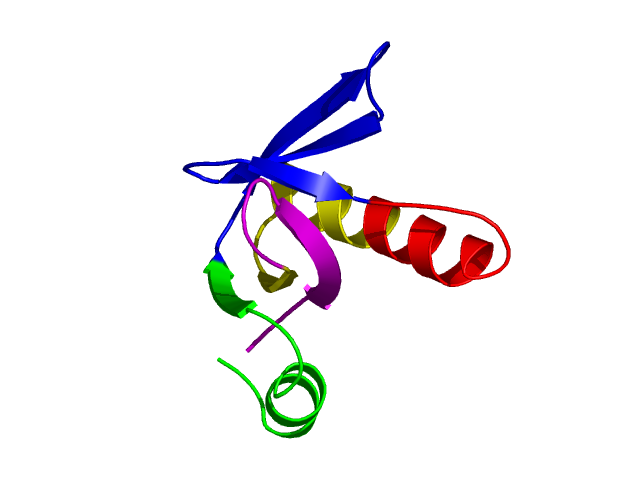
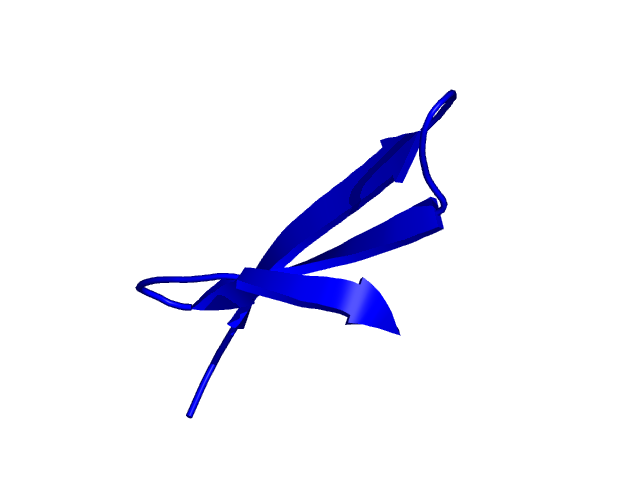
Parameters choosen
R-value: 95
Minimal size of secondary structure: 8
Minimal size of protein unit: 16
Maximal size of protein unit: 0
Pruning tree ? No
CI cut-off: 0.2
Cut-off distance between atom: 8
Delta value in logistic function: 1.5