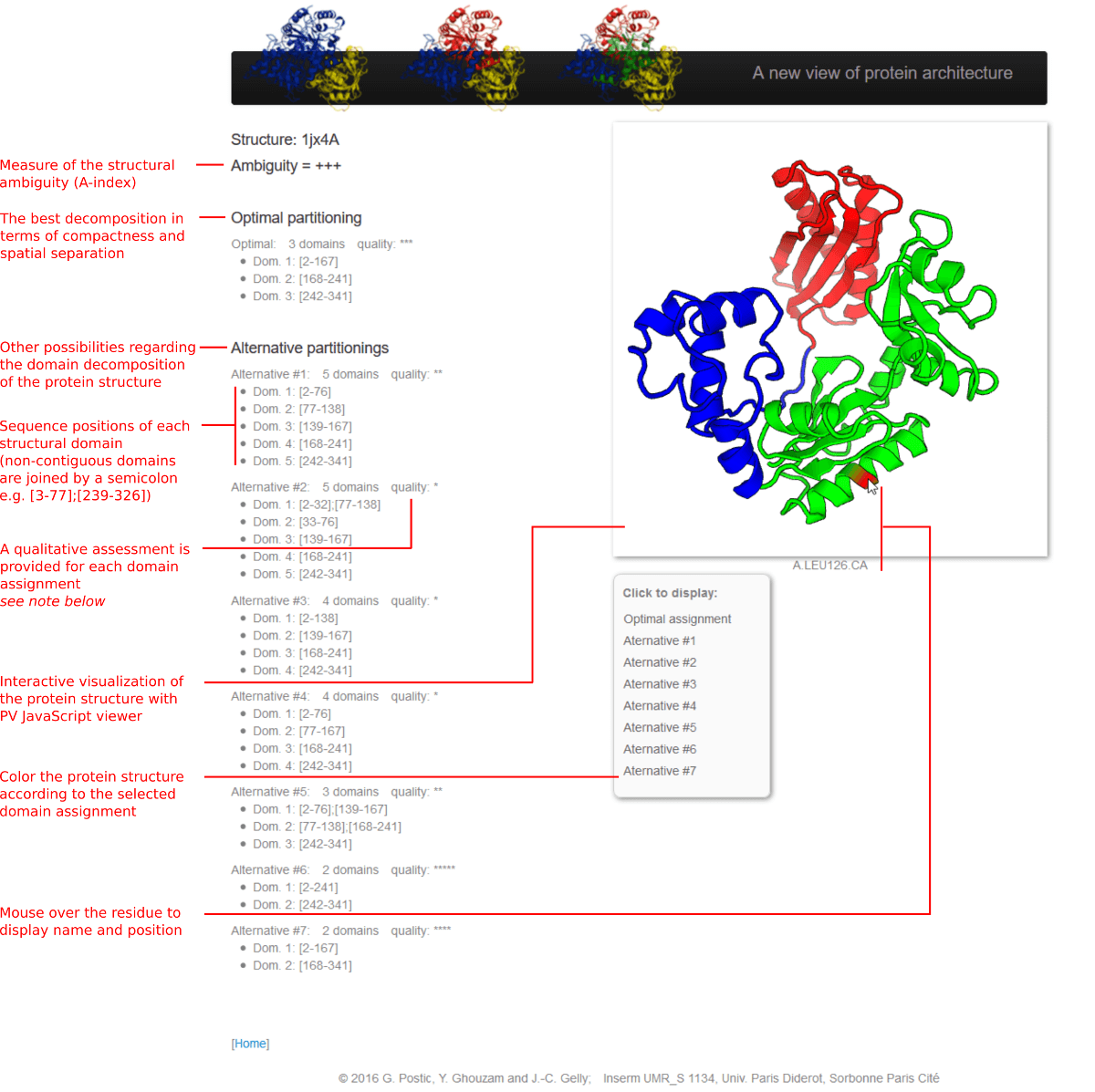
Note:
the quality corresponds to the Euclidean distance between the decomposition and the threshold of the acceptance region
(see model).
It is discretized into five values, from '*' to '*****' (a quality of '***' corresponds to a decomposition that is close to the threshold, either inside or outside the acceptance region).
In the above example, the optimal assignment has '***', although there is an alternative with a quality of '*****'. This is because when several decompositions fall in the acceptance region, the one that goes the furthest in partitioning the protein structure (i.e. the one that delimits the highest number of domains) is selected as the optimal. In this example, the 3-domain assignment is selected over the 2-domain assignment.
If there are 3 or more decompositions, each with a quality of '***' or more, then the structural ambiguity will be '+++' (A-index = 3). If there is no decomposition (1-domain assignment), then the A-index is null.
For readability, the number of alternative domain assignments displayed by the web server is limited. The same decompositions (and those that may have been hidden) can be obtained with the standalone version of SWORD, by using the '--max 9' option.
[Back]
© 2016 Guillaume Postic, Yassine Ghouzam & Jean-Christophe Gelly; Inserm UMR_S 1134, Univ. Paris Diderot, Sorbonne Paris Cité