Help
Search a variant
To search for a variant please go to the "Search" page.
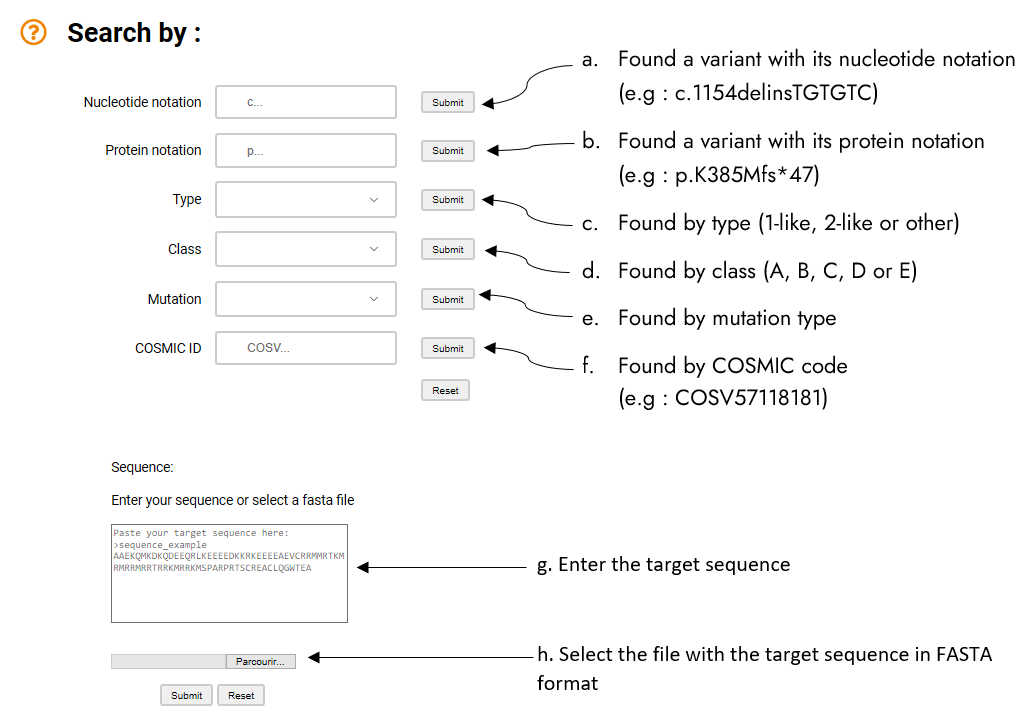
Seven independent options exist to search for a specific variants (see here for the complete liste).
Information on (a) the nucleotide notation (explanation), (b) the protein notation (explanation), or (f) the COSMIC code (explanation) can be entered. This information can be complete or partial, i.e for the protein, the request 385 will provide all the variants associated to this position.
Each variant is associated to different classifications, some are classical with the (type 1-like, type 2-like and others) or the mutation (frameshift, deletion, …); they can be selected with (c)
type or (d) mutation. We have developed a novel classification per classes from A to E (e) (explanation).
For more informations on these entries, please refer to the "Method" page, "Variant description" section.
A search can also be performed from a protein sequence. (f) The target sequence is pasted in FASTA format or plain text; a file in FASTA format can also be uploaded (only the first sequence will be use if multiple sequences are provided).
Only one entry can be used for a query.
Results found

The results appear in a new page, in a table. The information given for each variant is :(i) its nucleotide notation (nucleotide ID), (ii) protein notation (protein ID), (iii) its type, (iv) its class, (v) its mutation type and (vi) if avalaible its COSMIC code. The complete description page for a variant is accessible via its nucleotide notation.
Variant description
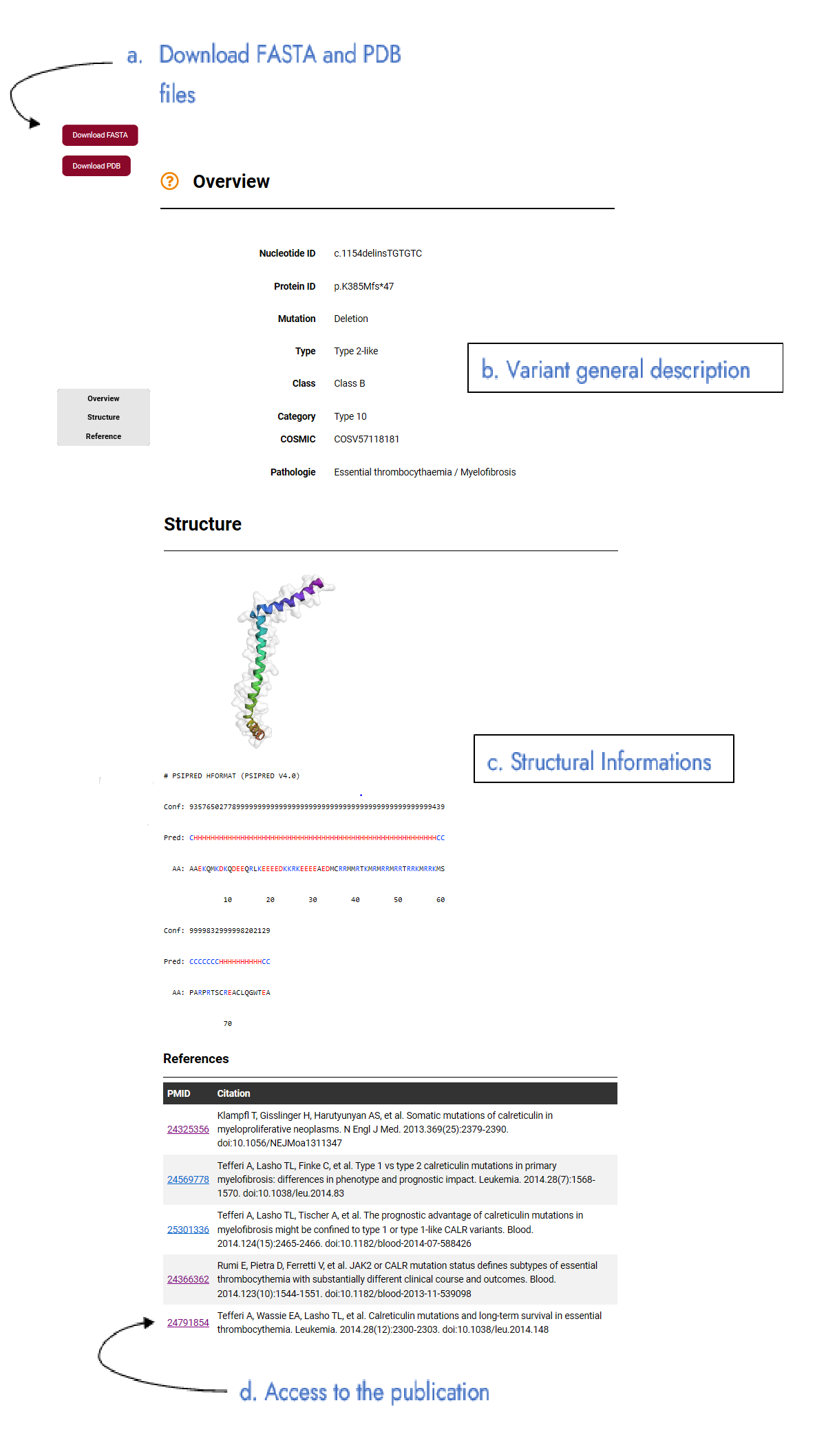
The description page of a variant is divided into 3 parts: - Overview. (b) It corresponds to the general description of the variant with (i) its nucleotide notation (nucleotide ID), (ii) protein notation (protein ID), (iii) its type, (iv) its class, (v) its mutation type, (vi) its categorization, (vii) if available its COSMIC code, and (viii) different associated pathologies. More information in "Method" page, "Variant description" section. - Structure. (c) Information on the secondary structure prediction with PSIPRED prediction and its confidence index are provided, as well as the best available structural model structure of a variant. For the latter, the 3Dmol.js applet is used and can be used interactively. More information in "Method" page, "Structure data" section. - References. (d) All variant references with access to the article via PMID are provided. The sequence in FASTA format and the 3D model in PDB format can be downloaded via the buttons (a) at the top left of the page.
No result found
If no results were found, the type and the class of the target sequence can still be determined. To perform this task, the variant sequence can be either (i) whole with signal peptide (starting with residue M1), (ii) without the signal peptide (starting with residue E18), or (iii) give only the sequence of the C-domain from residue A352.