ORION principle
ORION (Optimized protein fold RecognitION) is a new profile-profile fold recognition approach that relies on a better description of the local protein structure to boost distantly protein detection. These descriptors called Protein Blocks (PB) encodes a structural alphabet defined by 16 folding patterns (see 1) that describe accurately local protein structures (de Brevern et al. Proteins 2000) contrary to the secondary structures which are composed of only 3 predicted states (helix , strand and coil). Sequence profile and predicted PB profile of target are combined to search by a profile-profile dynamic programming algorithm in a library of templates profiles build from structural alignments.
1 - Protein Blocks
ORION method
ORION is based on the pairwise comparison of profiles combining sequence and structural information.
The first step is the generation of the template profile library (see 2 - step A). Sequence profiles are obtained from multiple sequence alignments (MSA)
of HOMSTRAD families (Mizuguchi et al. Protein Sci. 1998) and represent the 20 amino acid scores for each position of the MSA. Structural profiles represent the 16 PB scores
for each position of the MSA. Templates structural MSAs were translated into PB MSAs using the atomic coordinates of the protein structures
to generate template PB profiles. Amino acid and PB profiles are joined to form 'AAPB profiles' containing both sequence and structural
evolutionary information.
From the query sequence, the second step consists in generating a query profile (see 2 - step B).
The query sequence profile is obtained after three iterations of PSI-BLAST and is used to predict the query PB profile.
The third and last step is the search for related proteins in the template profile library (see 2 - step C).
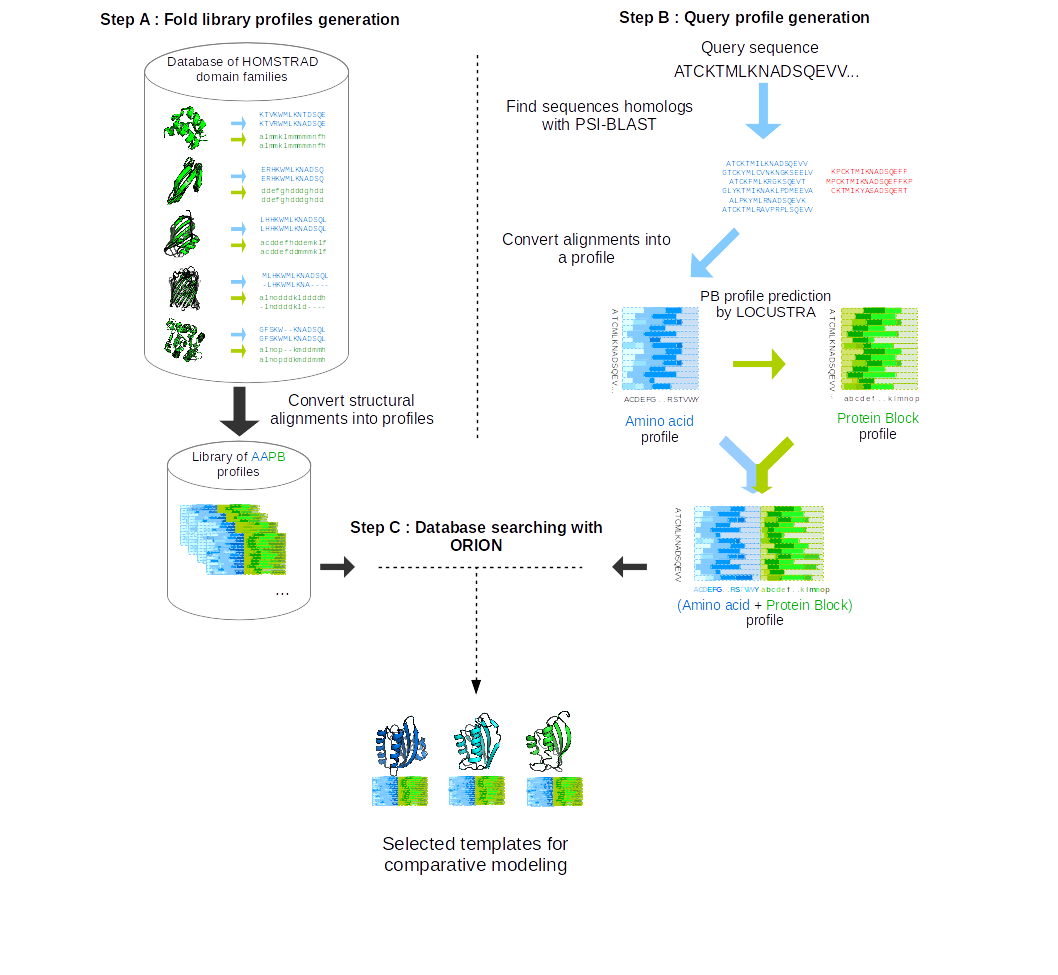
2 - ORION flowchart (from Ghouzam et al. Bioinformatics 2015)
ORION Performances
Identification of related proteins in CASP
Dataset : 150 template-based modeling targets (CASP8 + CASP9 + CASP10) with at least one template in the HOMSTRAD database with the same fold.
Method : ROC curve at fold level (See 3A) (True positive rate (TPR) vs False positive rate (FPR)) , Success rate at fold level for top 1, 5 and top 10 (See 3B).
Conclusion : At 10% of FPR ORION performs with 40% of TPR, 2x more than HHsearch and 2.5x more than PSI-BLAST
ORION detects 8.6% more homologs than HHsearch and 22.9% more than PSI-BLAST at top 10.
ORION performs better than HHsearch and PSI-BLAST in the detection of CASP templates
