Main goal
In recent years the number of observed protein folds and the number of available structural data have hugely increased (respectively 1194 and 88170 on February 2013).
This emergence of data became extremely helpful for different research fields as molecular modeling and phylogeny. Indeed, comparison and classification of protein structures provide evolutionary relationships between proteins.
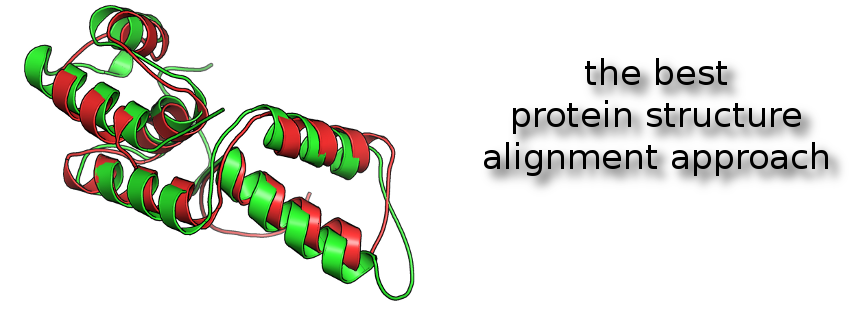
One of powerful way to compare proteins and detect structural analogies is the superimposition of protein structures.
Principle
iPBA methodology (1,2) was benchmarked as the best actual approach to superimpose 3D protein structures. It's based on a structural alphabet, named the Protein Blocks (3,4).
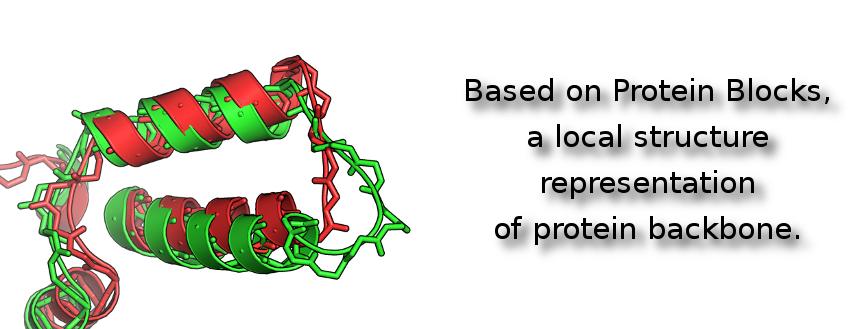
The Protein Blocks allow to represent the 3D structure of proteins as a sequence of letters (blocks). Thereby the problem of protein structural comparison is reduce to classical sequence alignment.
It's known that protein structures are more conserved than amino-acid sequences.
As the Protein Blocks represent pentapeptide conformations of proteins backbones, this approach allows a structural comparison independent of the proteins amino-acid sequences.
Results
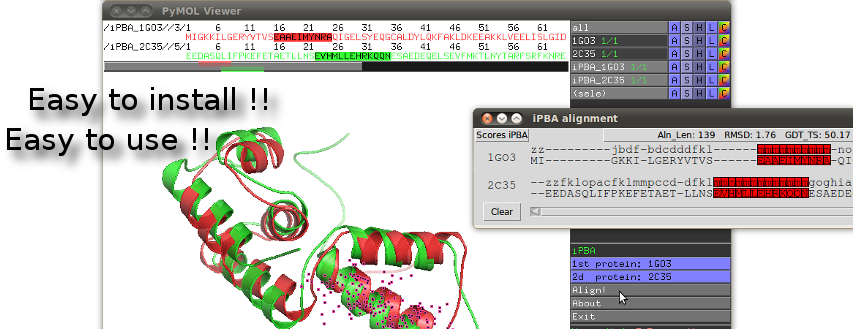
We proposed here, iPBAVizu, a plugin for the very popular molecular graphics viewer PyMOL (5).
Easy to install and easy to use, this plugin is a simple way to visualize and appreciate the iPBA methodology.
Through his interactive window, iPBAVizu gives several alignment quality measurement (such like GDT_TS, RMSD ...), and provides a simple way to explore alignments in terms of amino-acid sequence, Protein Blocks, and 3D protein structures.
In summary, with iPBAVizu, you have access to the best protein structure alignment approach into the most widely used protein visualisation tool.
Enjoy your proteins superimpositions!!!
References
- Joseph AP, Srinivasan N, de Brevern AG (2011) Improvement of protein structure comparison using a structural alphabet. Biochimie
[article] - Gelly JC, Joseph AP, Srinivasan N, de Brevern AG (2011) iPBA: a tool for protein structure comparison using sequence alignment strategies. Nucleic Acids Res.
[article] - de Brevern, AG, Etchebest, C, and Hazout, S (2000) Bayesian probabilistic approach for predicting backbone structures in terms of protein blocks. Proteins.
[article] - Joseph AP, Agarwal G, Mahajan S, Gelly J-C, Swapna LS, Offmann B, Cadet F, Bornot A, Tyagi M, Valadie´ H, Schneider B, Cadet F, Srinivasan N, de Brevern AG (2010) A short survey on protein blocks. Biophys Rev.
[article] - DeLano WLT (2002) The PyMOL molecular graphics system. Schrodinger, Delano Scientific LLC, San Carlos.
[website]
If you use iPBAVizu for a publication, please cite it!
........... Structural Bioinformatics ..................