
Introduction
MAIDEN (Model quality Assessment for Intramembrane Domains using an ENergy criterion) is a statistical potential optimized on native alpha-helica and beta-sheet membrane protein structures. MAIDEN computes pseudo-energies for transmembrane domains of native or predicted protein structures, in order to evaluate their quality.
Reference:
Download MAIDEN
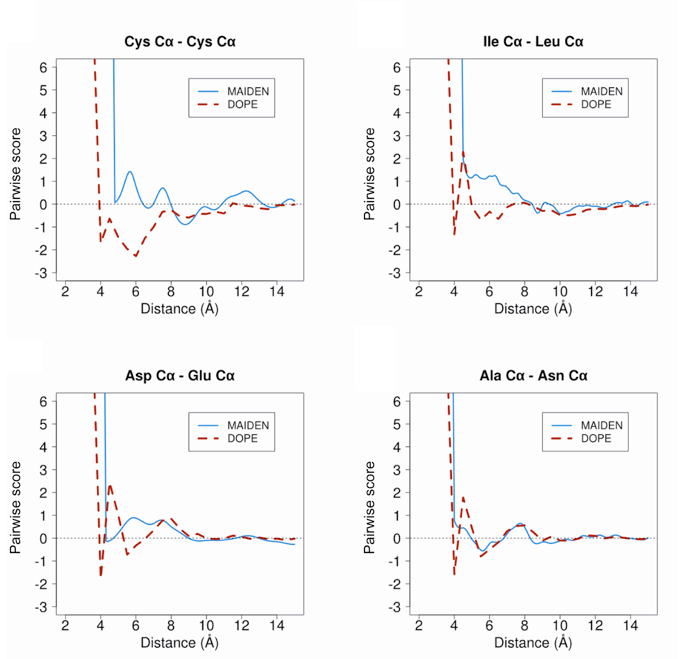
Training set
The set of structures used for the calculation of MAIDEN contains 66 representative structures (41 alpha and 25 beta) from the PDBTM database (Tusnady, et al., 2005) determined by crystallography at <2.5 A resolution and with an R-factor <0.3 (see below):
1JB0 2F93 2VDF 3EMN 3SZV 4AL0 1KQF 2FGQ 2W2E 3GIA 3TDS 4AMJ 1OKC 2GR7 2WJR 3GP6 3TIJ 4DVE 1QD5 2HDI 2WSW 3HD6 3TX3 4DX5 1QJP 2J58 2X27 3KVN 3V5U 4E1S 1U19 2J7A 2X55 3M73 3VY8 4EIY 1UUN 2MPR 2X9K 3N5K 3W54 4ENE 1XKW 2O4V 2XOV 3PCV 3WBN 4EZC 1YC9 2Q67 2YEV 3PGU 3ZUX 4HFI 2A65 2QTS 3ARC 3QRA 4A01 4IKW 2BL2 2R9R 3DH4 3RLF 4AFK 7AHL
These representative structures share no more than 30% sequence identity with each other. The list was culled by entries from 1709 PDBTM structure files, using the PISCES server (Wang and Dunbrack, 2003; Wang and Dunbrack, 2005). Fold conservation in transmembrane regions requires less sequence identity than for water-soluble proteins (Olivella, et al., 2013). Therefore, even when sharing <30% sequence identity, proteins belonging to the same structural family based on (Lomize, et al., 2012) were also filtered, making our training set non-redundant both in terms of sequence identity and structure similarity. The assignments of intramembrane residues were obtained using the TMDET web server (Tusnady, et al., 2005).

Models and detailed results
To benchmark MAIDEN and others methods, we have generated 700 predicted structures (alpha and beta) by homology modelling and used the 15340 membrane protein models of EVfold_membrane (Hopf et al., 2012).
Download models and detailed results
References
- Hopf, T.A., et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell 2012;149(7):1607-1621.
- Lomize, M.A., et al. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic acids research 2012;40(Database issue):D370-376.
- Olivella, M., et al. Relation between sequence and structure in membrane proteins. Bioinformatics 2013;29(13):1589-1592.
- Tusnady, G.E., Dosztanyi, Z. and Simon, I. TMDET: Web server for detecting transmembrane regions of proteins by using their 3D coordinates. Bioinformatics 2005;21:1276-1277.
- Wang, G. and Dunbrack, R.L., Jr. PISCES: recent improvements to a PDB sequence culling server. Nucleic acids research 2005;33(Web Server issue):W94-98.
- Wang, Z., Tegge, A.N. and Cheng, J. Evaluating the absolute quality of a single protein model using structural features and support vector machines. Proteins 2009;75(3):638-647.
- Postic G, Ghouzam Y, Gelly JC. An empirical energy function for structural assessment of protein transmembrane domains. Biochimie. 2015 Aug;115:155-61.