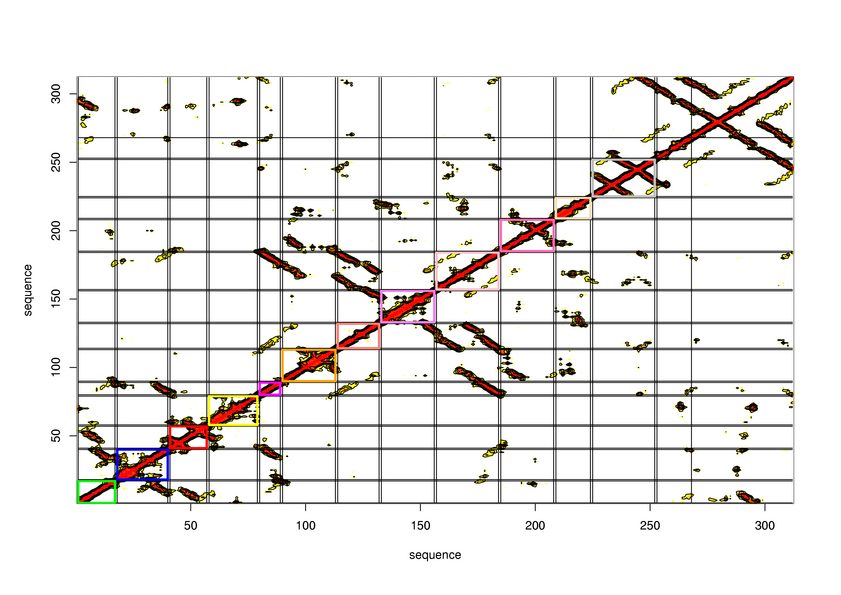
Parameters choosen
R-value: 95
Minimal size of secondary structure: 8
Minimal size of protein unit: 16
Maximal size of protein unit: 0
Pruning tree ? No
CI cut-off: 0.2
Cut-off distance between atom: 8
Delta value in logistic function: 1.5
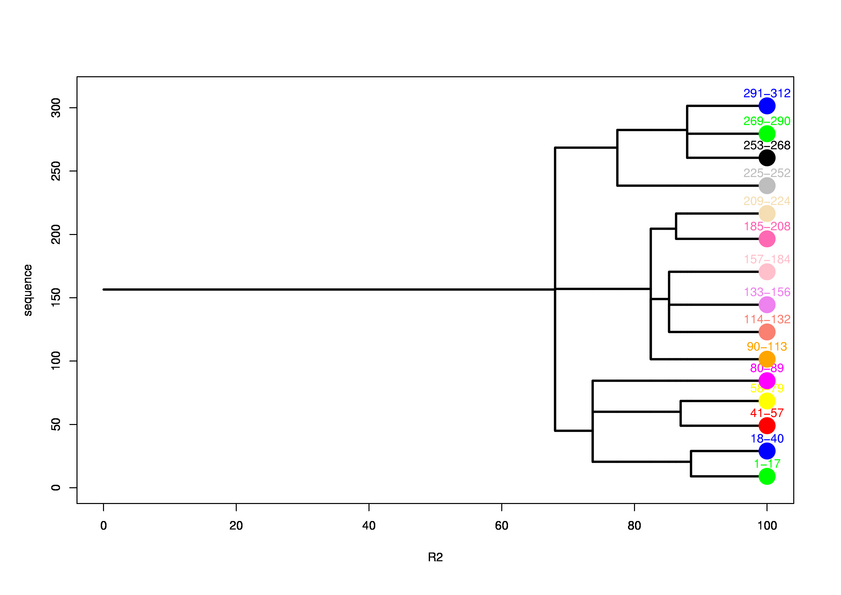
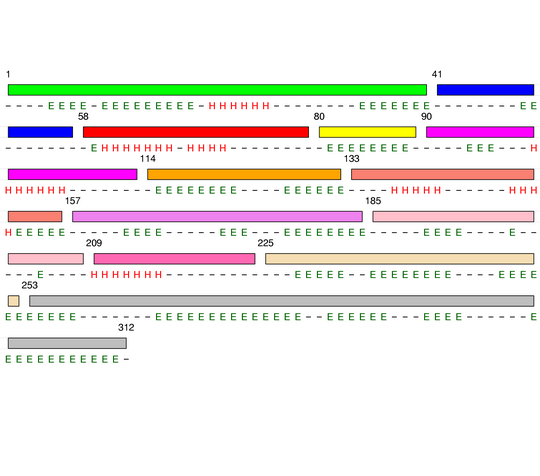
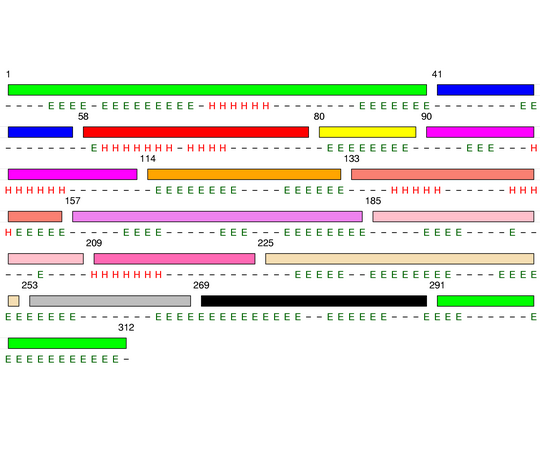
Hierarchical process of splitting
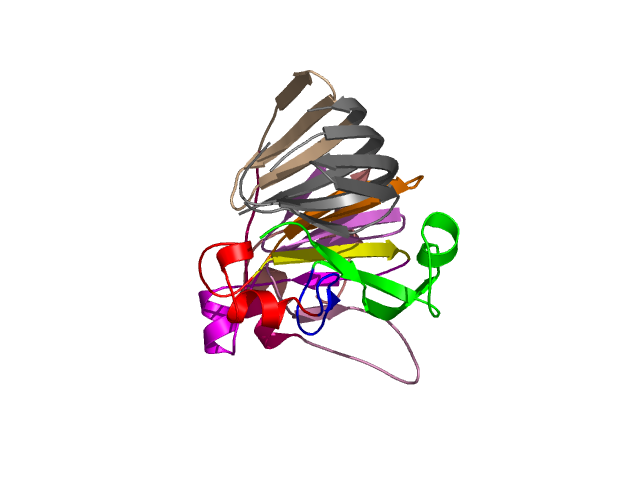
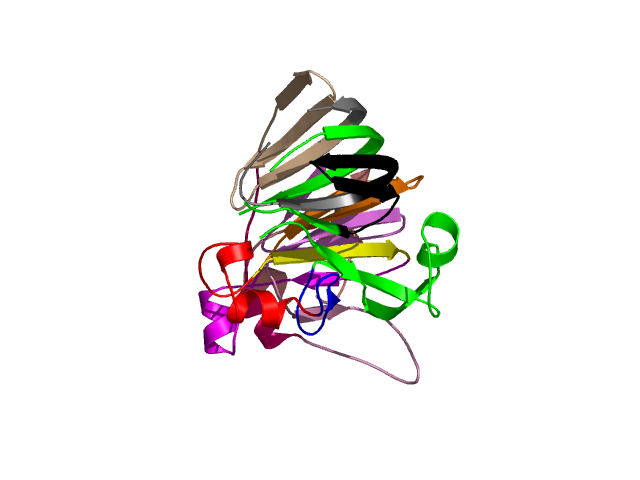
Visualisation of Protein units at all levels
Level 0

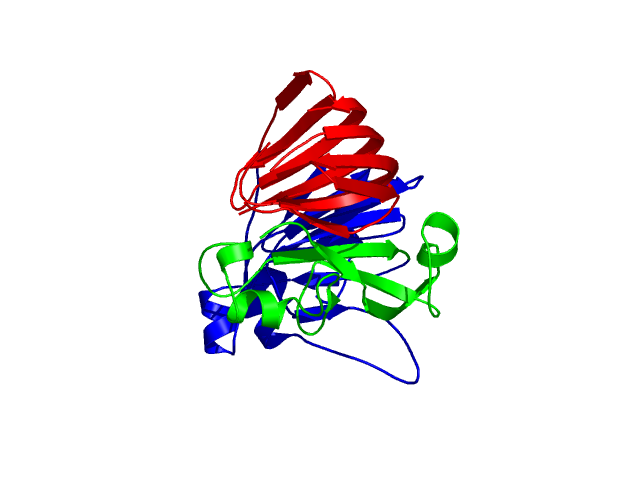
[1-89]:
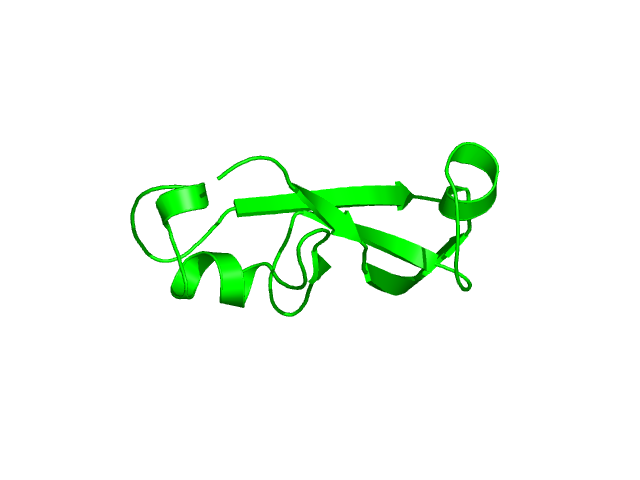
CI=4.00
[90-224]:

CI=4.62
[225-312
]:
CI=5.22
Level 1

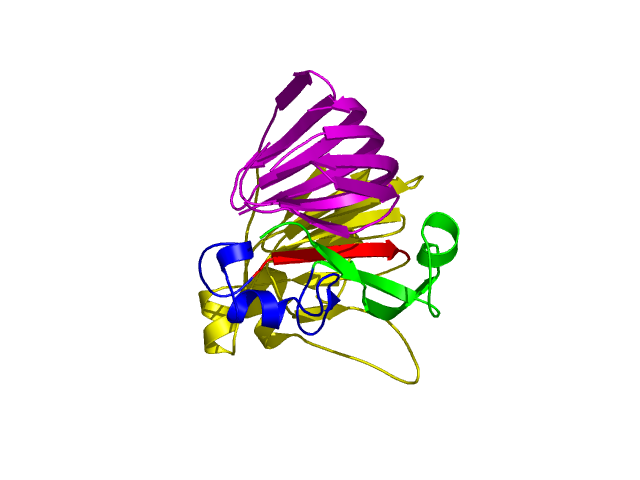
[1-40]:

CI=2.82
[41-79]:

CI=1.90
[80-89]:
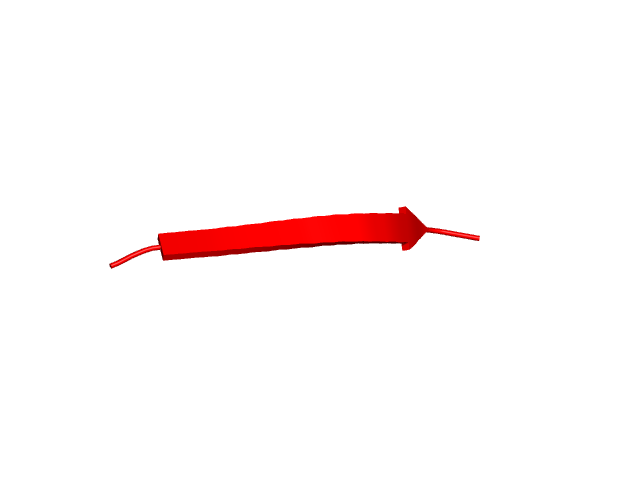
CI=0.00
[90-224]:
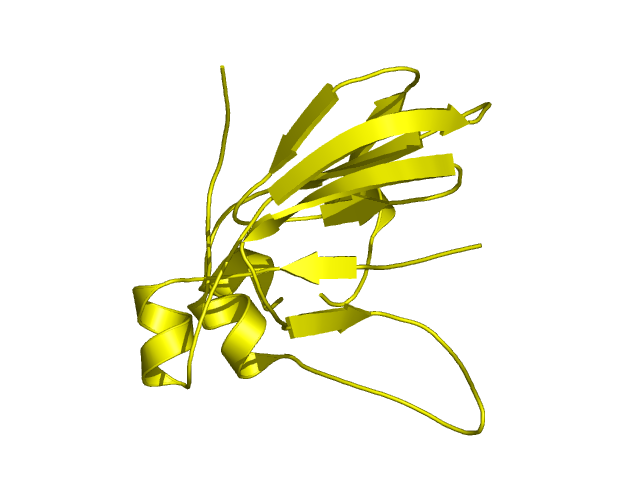
CI=4.62
[225-312
]:
CI=5.22
Level 2
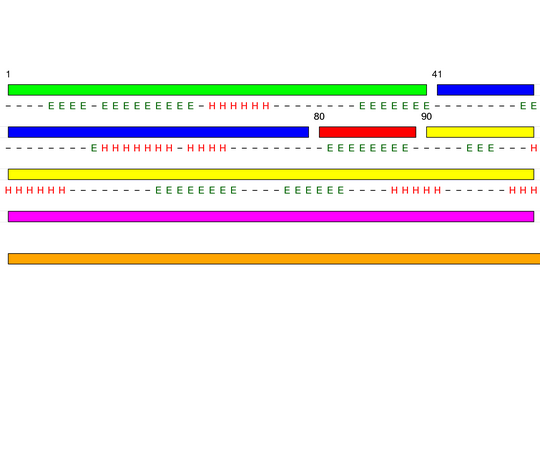
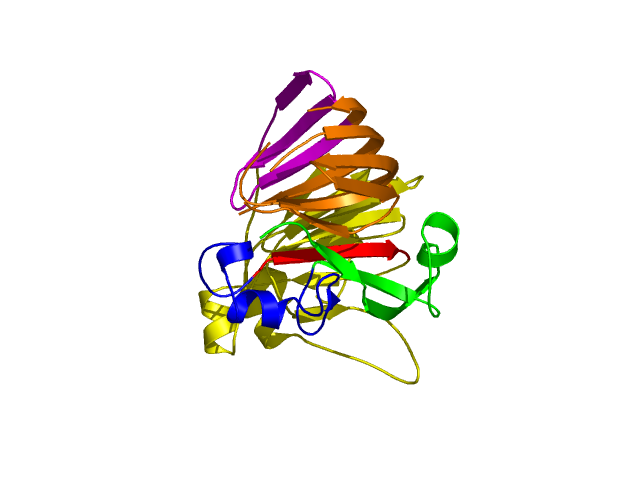
[1-40]:

CI=2.82
[41-79]:

CI=1.90
[80-89]:
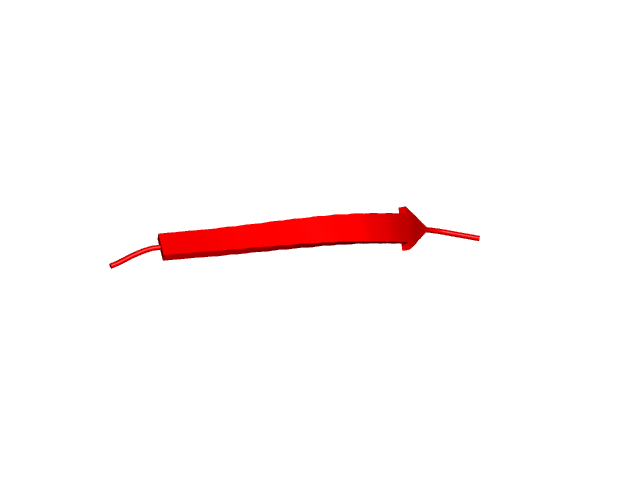
CI=0.00
[90-224]:
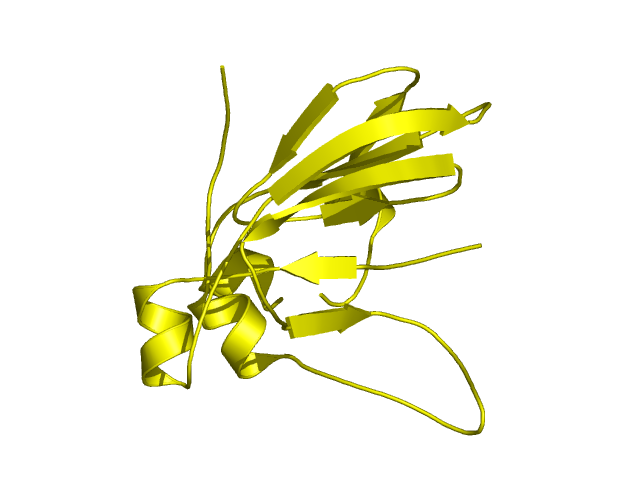
CI=4.62
[225-252]:

CI=3.12
[253-312
]:
CI=4.78
Level 3

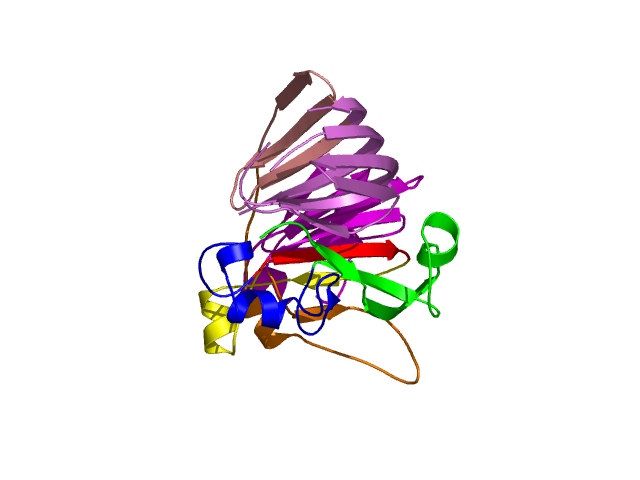
[1-40]:

CI=2.82
[41-79]:

CI=1.90
[80-89]:
CI=0.00
[90-113]:
CI=1.65
[114-184]:

CI=4.01
[185-224]:

CI=1.24
[225-252]:

CI=3.12
[253-312
]:
CI=4.78
Level 4
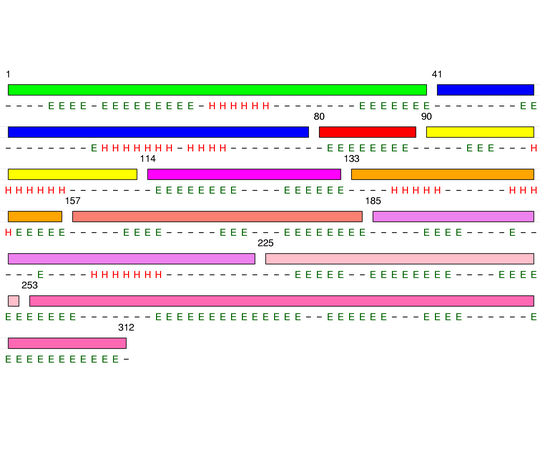
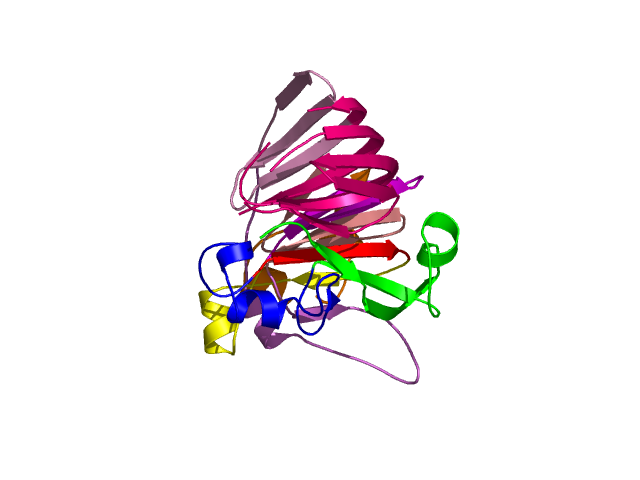
[1-40]:

CI=2.82
[41-79]:

CI=1.90
[80-89]:
CI=0.00
[90-113]:
CI=1.65
[114-132]:

CI=0.00
[133-156]:
CI=0.78
[157-184]:

CI=0.07
[185-224]:

CI=1.24
[225-252]:

CI=3.12
[253-312
]:
CI=4.78
Level 5
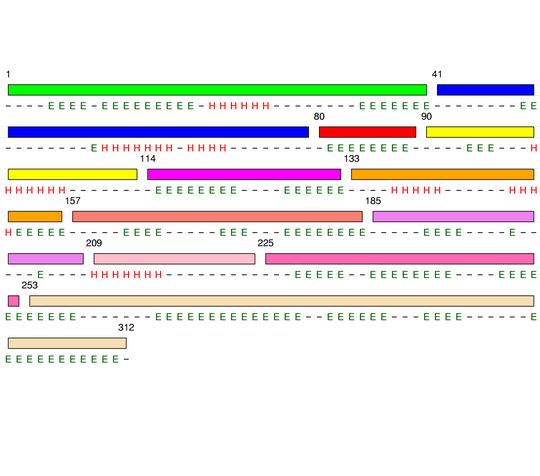
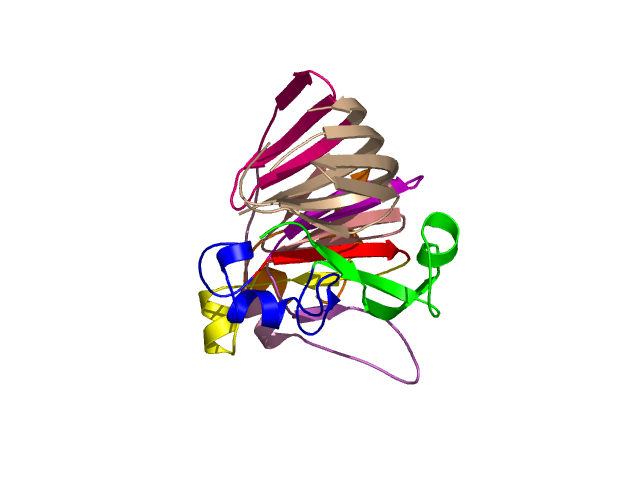
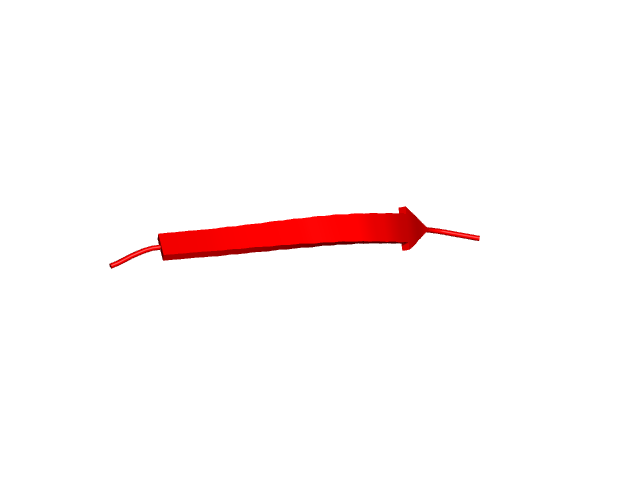
[1-40]:

CI=2.82
[41-79]:

CI=1.90
[80-89]:
CI=0.00
[90-113]:
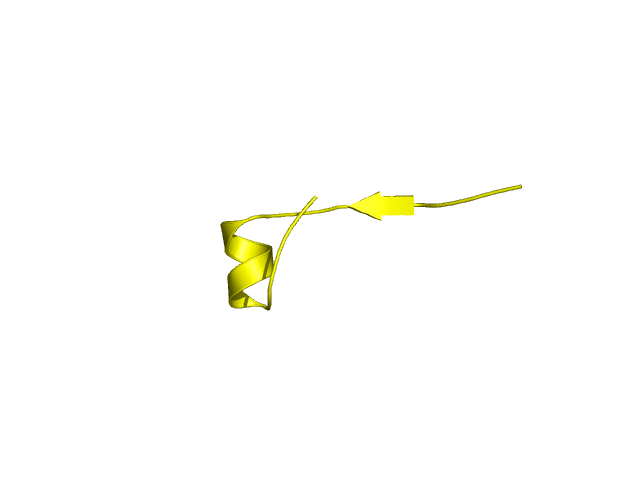
CI=1.65
[114-132]:

CI=0.00
[133-156]:
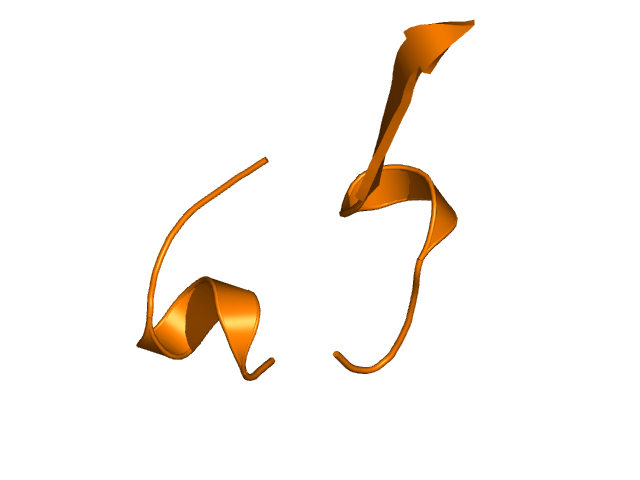
CI=0.78
[157-184]:

CI=0.07
[185-208]:
CI=1.40
[209-224]:

CI=0.00
[225-252]:

CI=3.12
[253-312
]:
CI=4.78
Level 6
[1-40]:

CI=2.82
[41-57]:

CI=0.88
[58-79]:

CI=0.75
[80-89]:

CI=0.00
[90-113]:
CI=1.65
[114-132]:

CI=0.00
[133-156]:
CI=0.78
[157-184]:

CI=0.07
[185-208]:

CI=1.40
[209-224]:

CI=0.00
[225-252]:

CI=3.12
[253-312
]:
CI=4.78
Level 7
[1-40]:

CI=2.82
[41-57]:

CI=0.88
[58-79]:

CI=0.75
[80-89]:

CI=0.00
[90-113]:
CI=1.65
[114-132]:

CI=0.00
[133-156]:
CI=0.78
[157-184]:

CI=0.07
[185-208]:

CI=1.40
[209-224]:

CI=0.00
[225-252]:

CI=3.12
[253-268]:
CI=0.00
[269-290]:

CI=2.58
[291-312
]:
CI=0.00
Final level 8
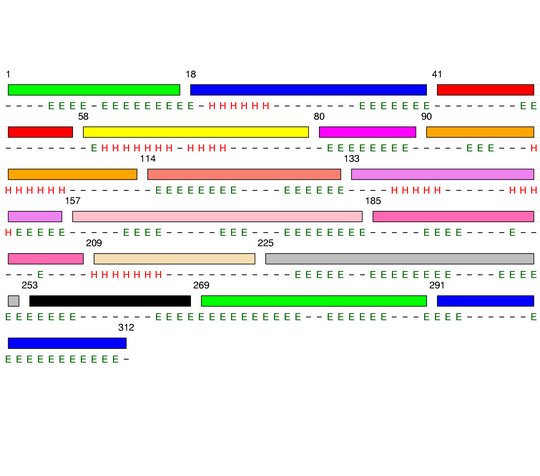
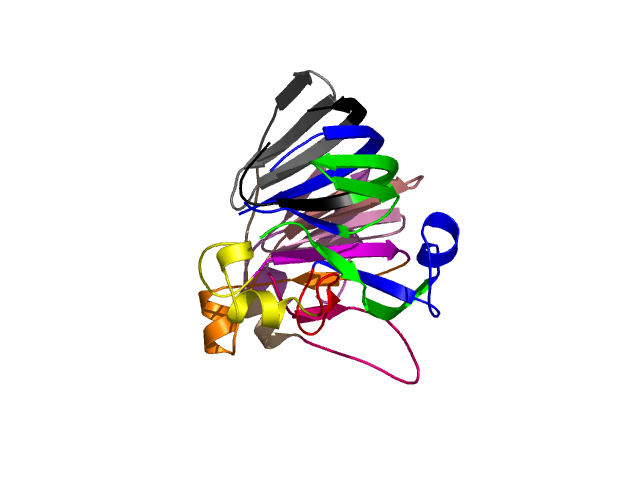
[1-17]:
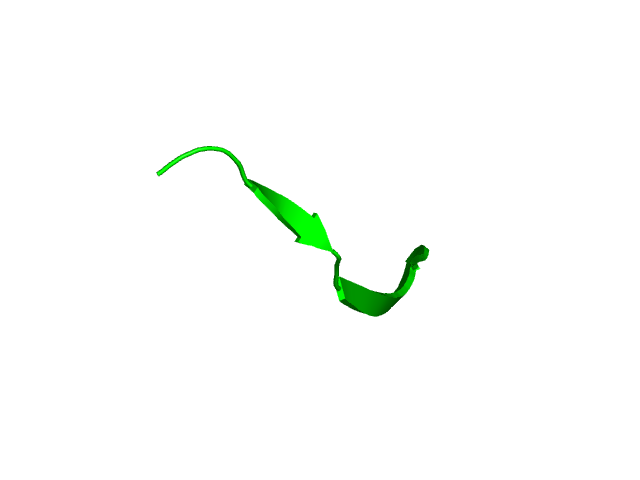
CI=0.00
[18-40]:

CI=1.38
[41-57]:

CI=0.88
[58-79]:

CI=0.75
[80-89]:

CI=0.00
[90-113]:

CI=1.65
[114-132]:

CI=0.00
[133-156]:
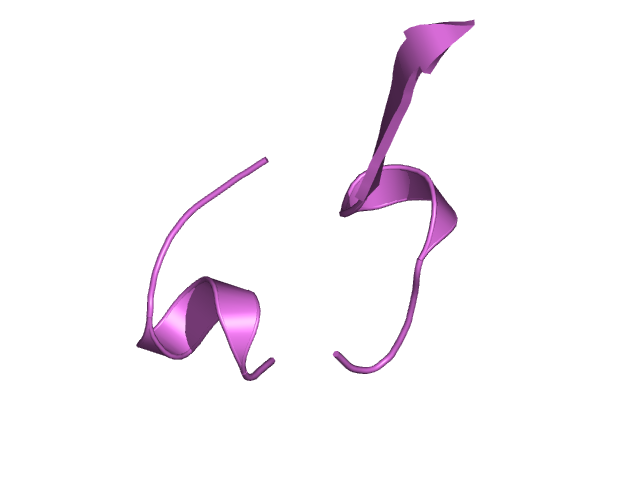
CI=0.78
[157-184]:

CI=0.07
[185-208]:
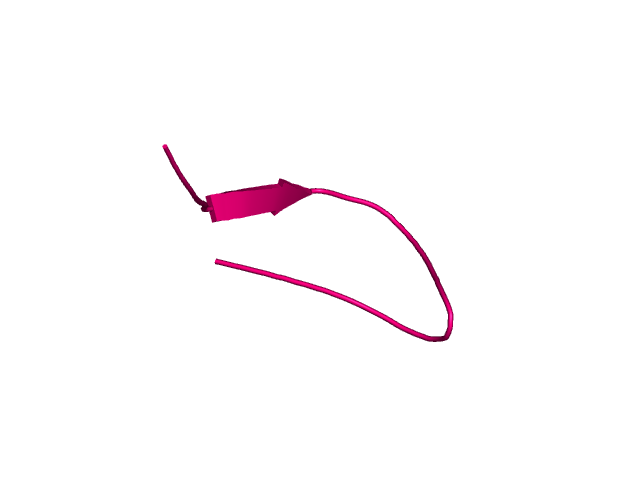
CI=1.40
[209-224]:

CI=0.00
[225-252]:

CI=3.12
[253-268]:
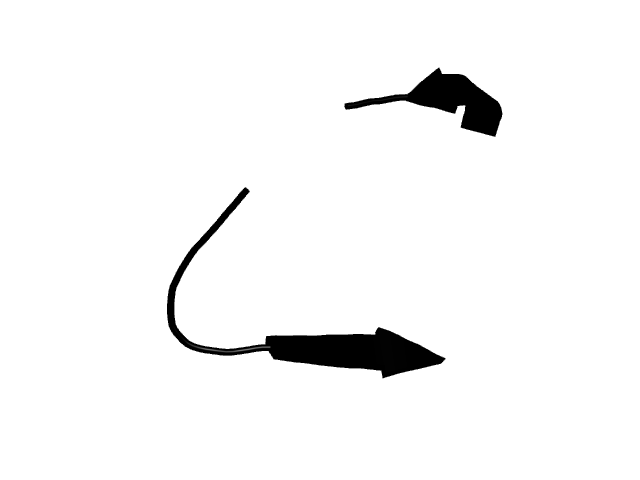
CI=0.00
[269-290]:

CI=2.58
[291-312
]:
CI=0.00