Aim
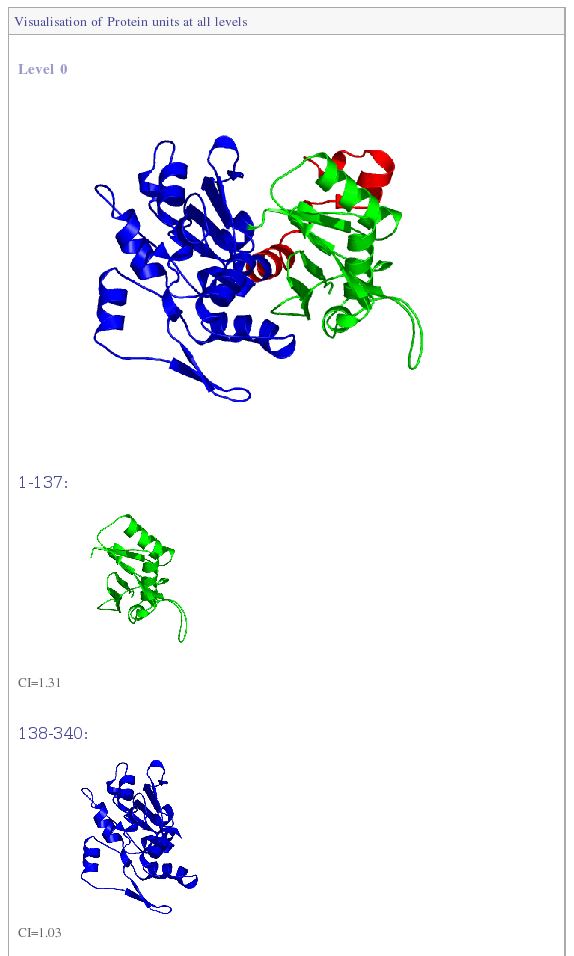
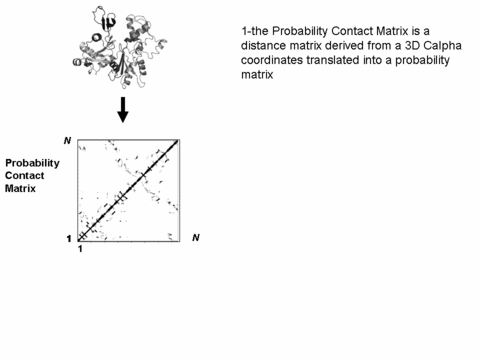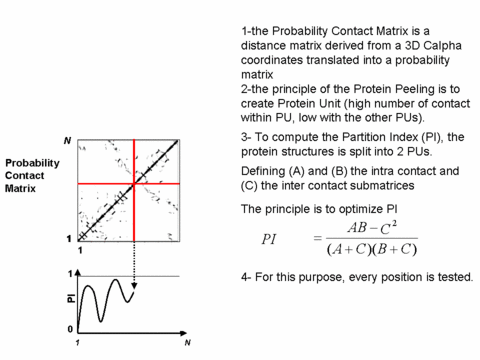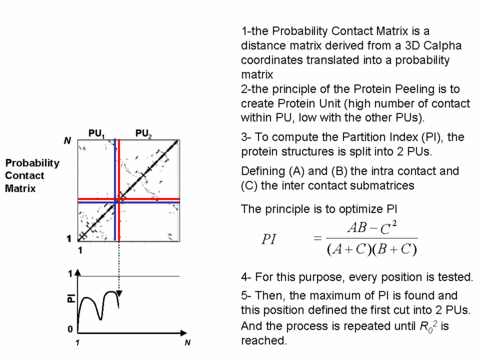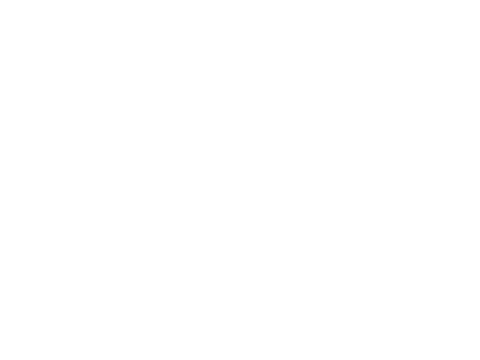
The approach "Protein Peeling" aims at cutting the 3D protein structure into a limited set of "Protein Units". A more precise definition of protein unit is a compact sub-region of the 3D structure corresponding to one sequence fragment. The basic rule lies upon the fact that for the considered protein fragment, the number of intra-PU contacts is the highest, and, the number of inter-PU contacts is the lowest.
The algorithm principle is to define a series of successive nested partitions; this leading to the building of a tree (or a hierarchy) showing the successive splitting of the PUs into sub-PUs.
Theoretically, protein units fill the gap between the concepts of secondary structure and structural domain, allows to link structural domain definition to secondary and super-secondary structure.
Illustrated principle
Tutorial
In this section we describe the principals ajustments and options to "peeling" a protein.
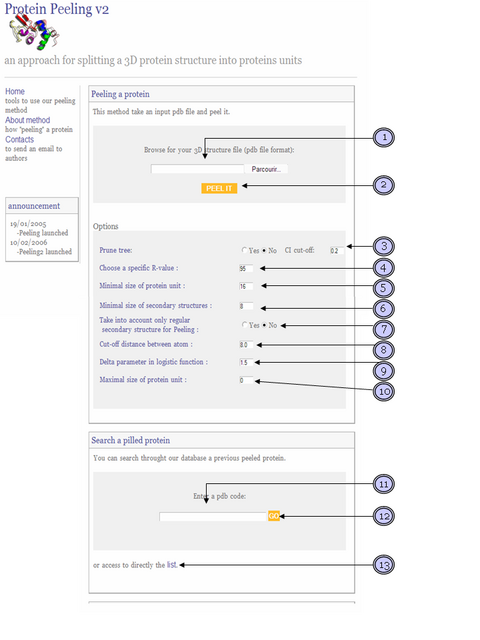
1) Browse your computer to retrieve your own three dimensional coordinates protein in pdb format (only one chain).
2) After path submited, click this button to begin the procedure. It can take up to 10 minutes, please be patient and don't refresh the page.
3) This parameter allow control the overall quality of splitting. With this parameter activated, program stop peeling if a splitting event produce only protein units (PUs) having a compaction index (CI) inferior to a given cut-off. CI is an entropy derived measure focuses on contacts intra PU vs extra PU. A weak value (<0.2) specifie a PU unit extended, a high value (>0.5)a unit of type compact.
4) Modify R value for increase or decrease the importance of splitting. A hight R value imply a hight level of splitting.
5) Specify the minimal size allowed to protein unit produced by algorithm.
6) Under this value, continuous secondary structure were not splitted.
7) With this parameter activated only contacts between regular secondary structure are taken into account in peeling process. Contribution of others contacts in computation of R-value and Partition Index are drastically reduced.
8) This value is the reference distance in Angstrom separating Calpha. This parameter D0 was used in logistic function to derive contact matrix into contact probability matrix used in peeling algrithm. High delfa value implied more distant contact taken into accounts to compute logistic fonction (see Gelly et al. 2006 for more explanation).
9) Delta parameter used in logistic function (see Gelly et al. 2006 for more explanation).
10) Maximal size of protein unit. By default this value is not used. If a value is choosen, protein peeling process continu while protein unit size is superior to this value
11) You can search a specific protein by is pdb code throught our precutting database. Enter only one or few first letters of pdb code display a list of protein that begin by this letters.
12) After enter a word, click this button to search.
13) You can access to precutted proteins directly by a list.
Example
In this section an example of protein peeling is detailled step by step for actin (pdb code: 1atnA)
1) Browse your directory for your input pdb file
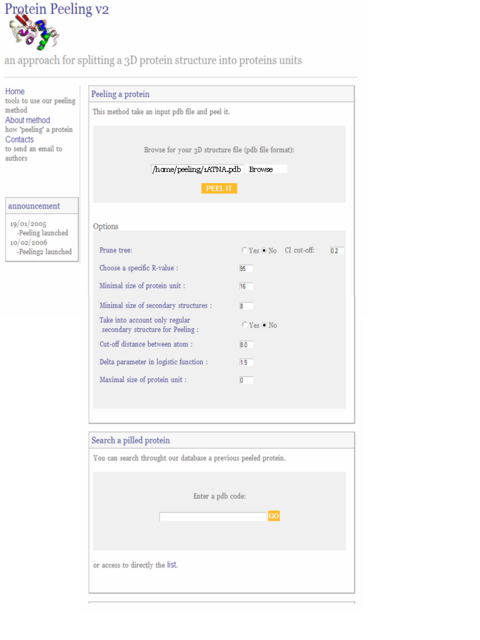
2) Lauch peeling process with "peel it" button and wait until results page is displayed
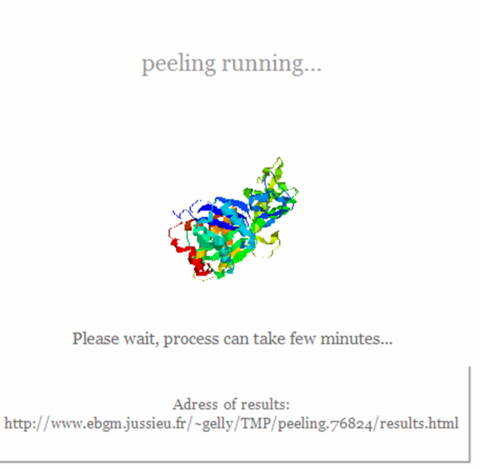
3) Results page is printed. Click on "GO" button to see 3D dimensionnal structure of protein unit produced.
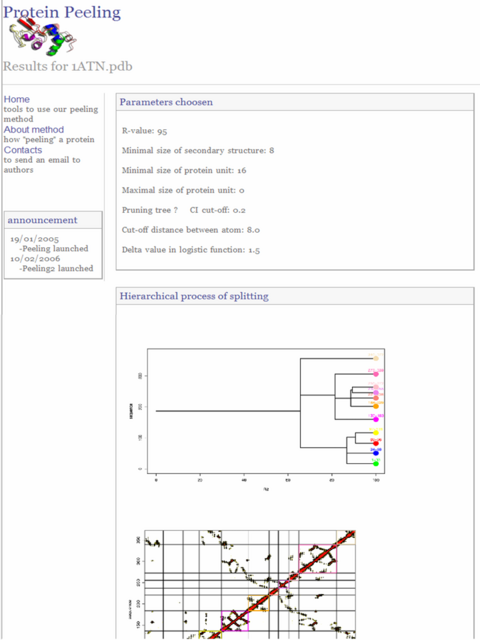
4) Wait until results page is displayed
5) New results page is printed. Click on "GO" button to see more detailled results for each level of peeling process.

6) Wait until results page is displayed
7) New results page with detailled process is printed.