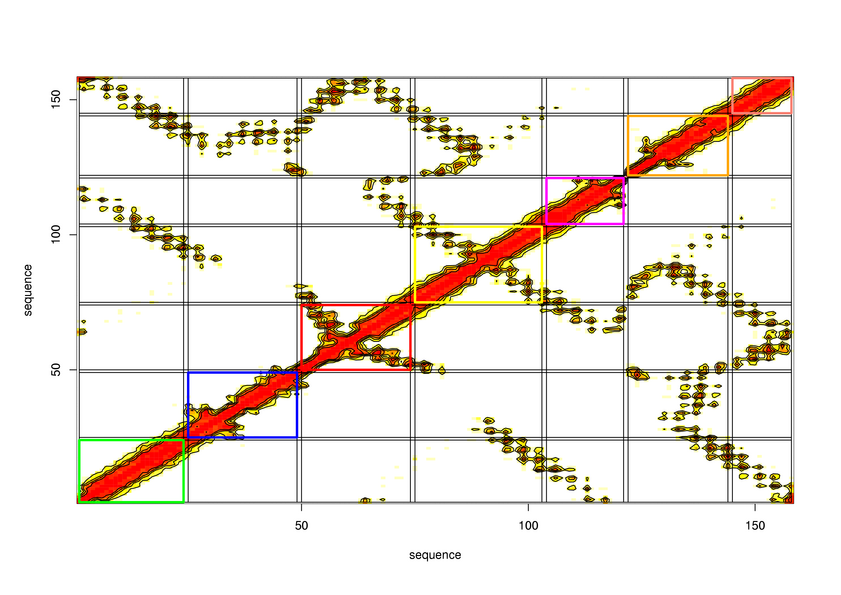
Parameters choosen
R-value: 95
Minimal size of secondary structure: 8
Minimal size of protein unit: 16
Maximal size of protein unit: 0
Pruning tree ? No
CI cut-off: 0.2
Cut-off distance between atom: 8
Delta value in logistic function: 1.5
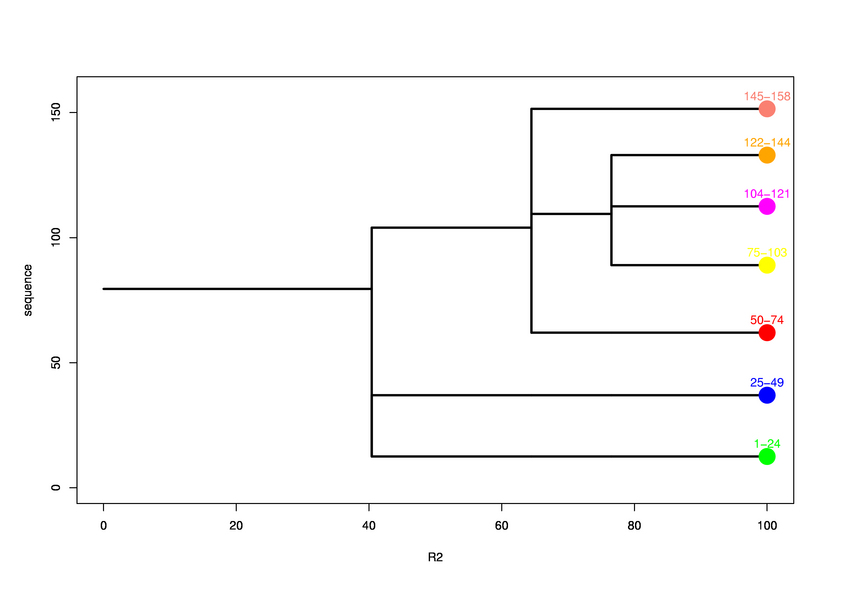
Hierarchical process of splitting
Visualisation of Protein units at all levels
Level 0
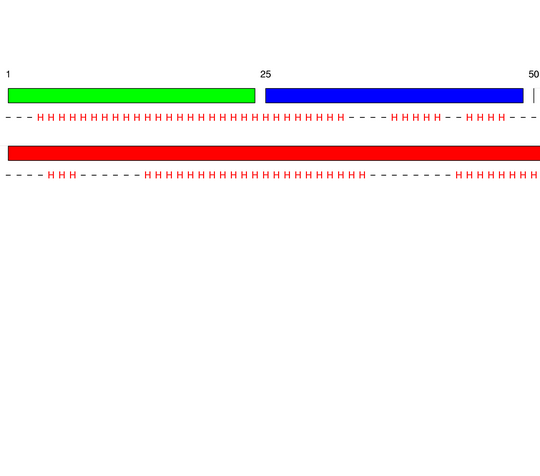
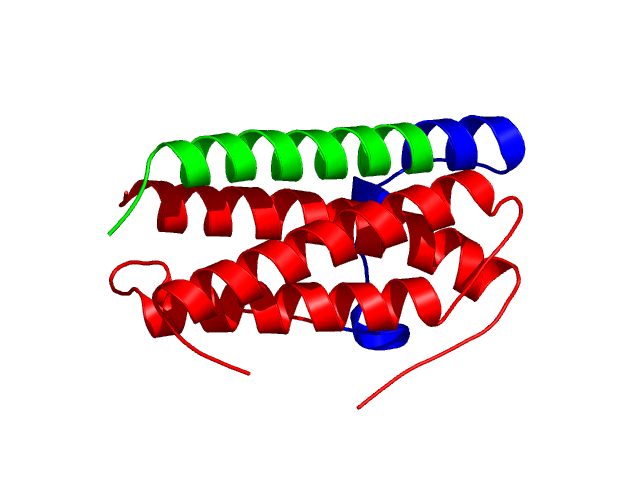
[1-24]:
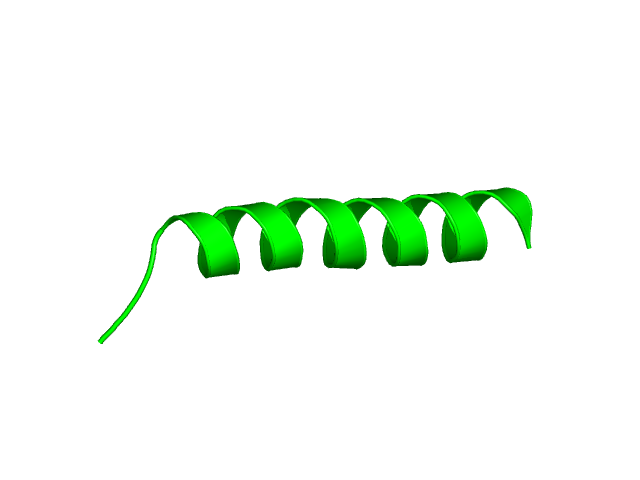
CI=0.07
[25-49]:
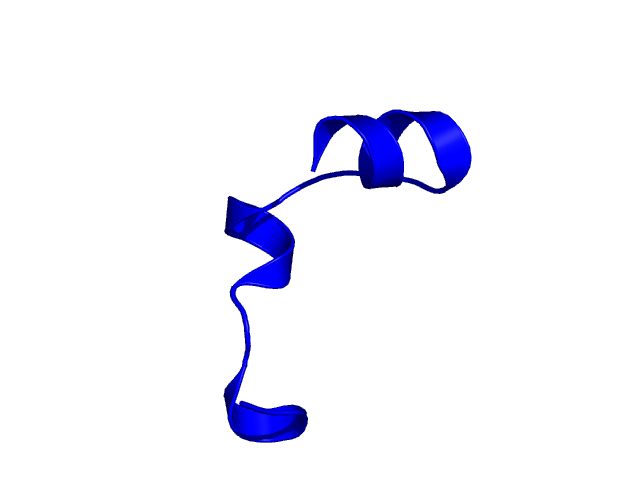
CI=0.52
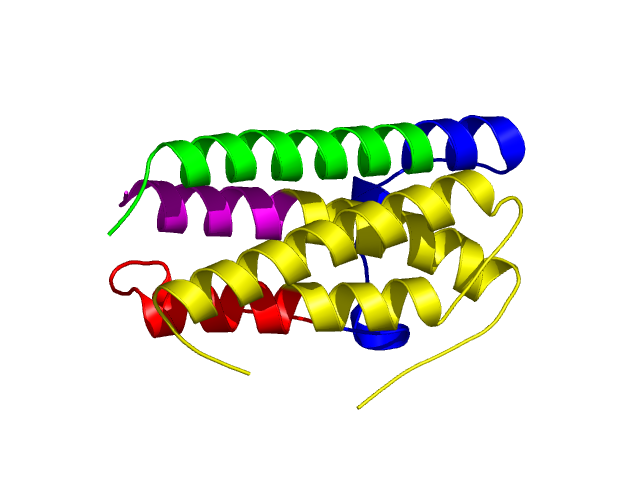
[50-158
]:
CI=2.63
Level 1
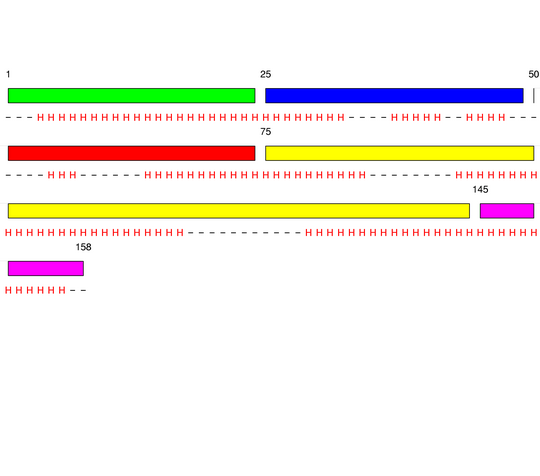
[1-24]:

CI=0.07
[25-49]:
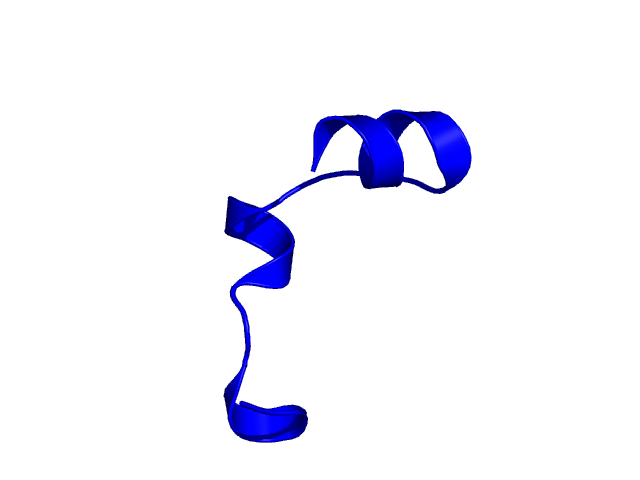
CI=0.52
[50-74]:

CI=2.53
[75-144]:
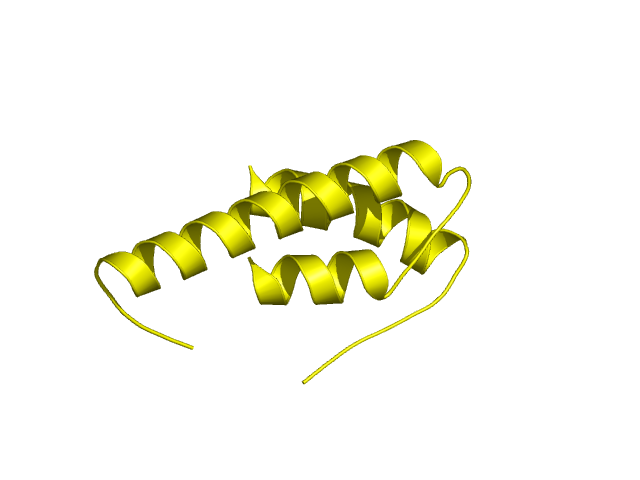
CI=1.57
[145-158
]:
CI=0.00
Final level 2
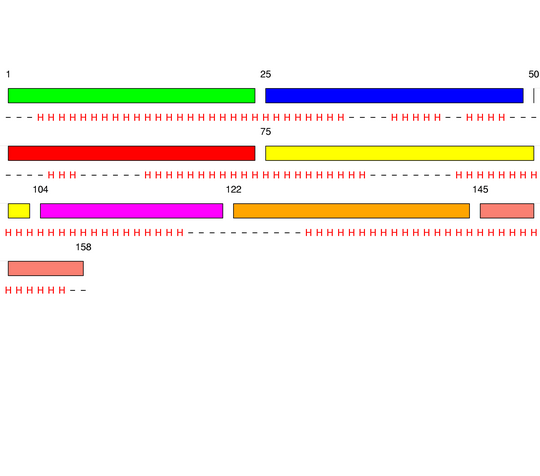
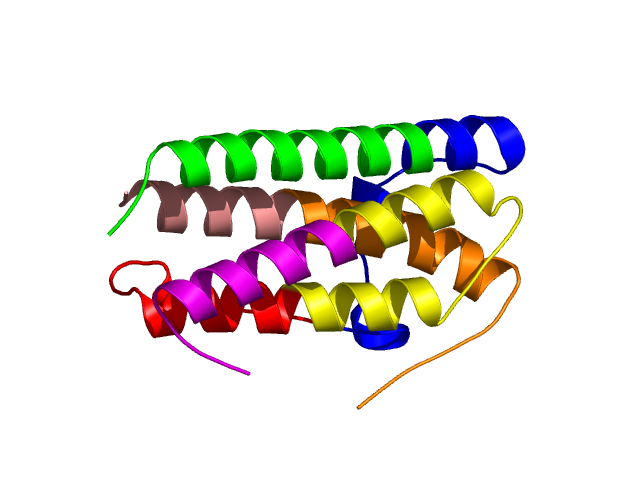
[1-24]:
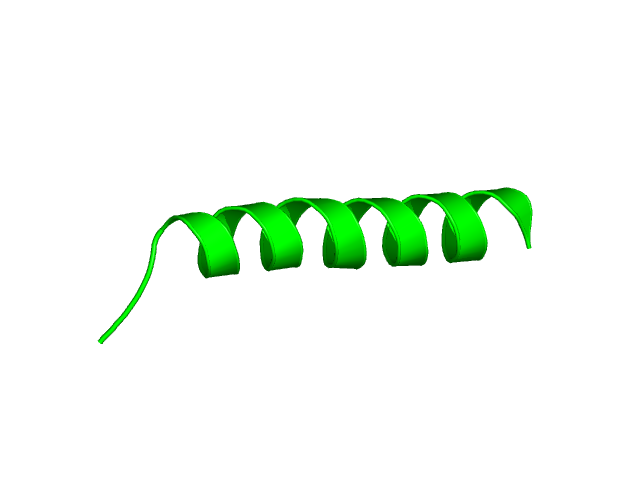
CI=0.07
[25-49]:
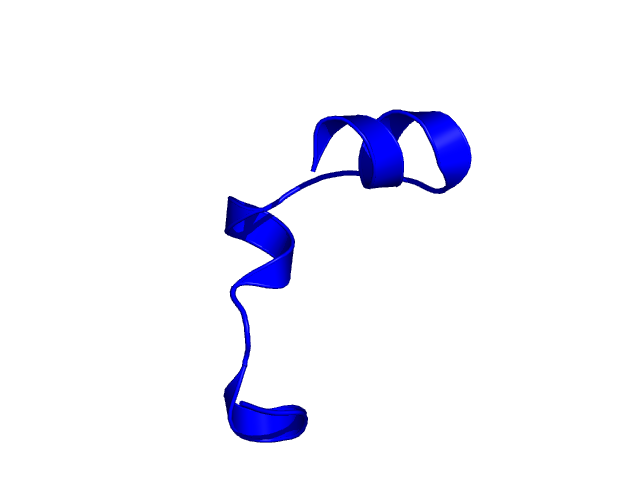
CI=0.52
[50-74]:

CI=2.53
[75-103]:
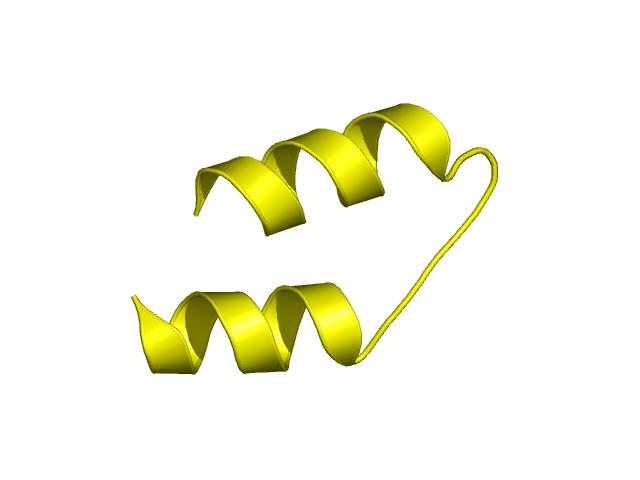
CI=1.09
[104-121]:

CI=0.31
[122-144]:

CI=0.08
[145-158
]:
CI=0.00