Parameters choosen
R-value: 95
Minimal size of secondary structure: 8
Minimal size of protein unit: 16
Maximal size of protein unit: 0
Pruning tree ? No
CI cut-off: 0.2
Cut-off distance between atom: 8
Delta value in logistic function: 1.5
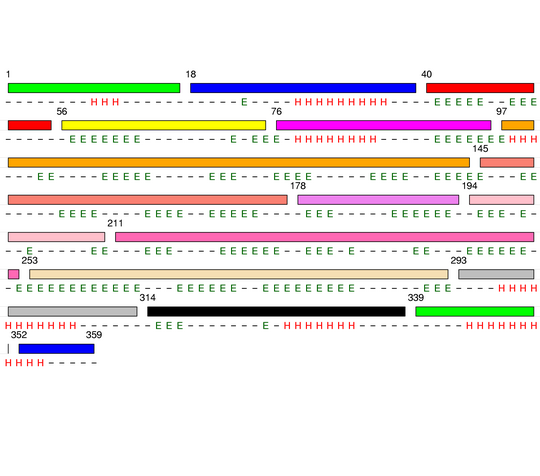
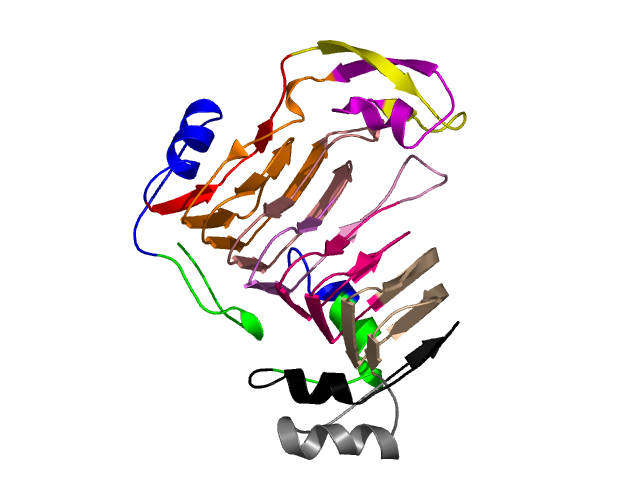
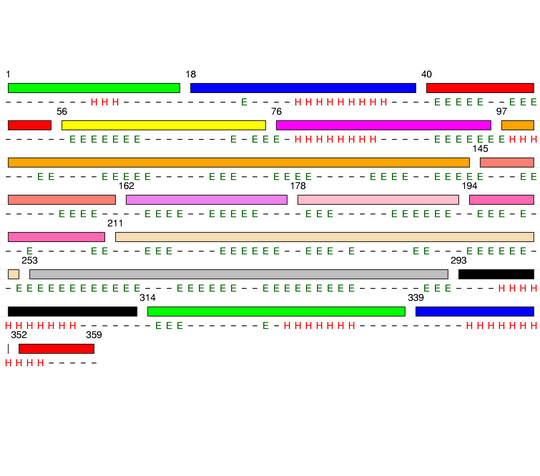
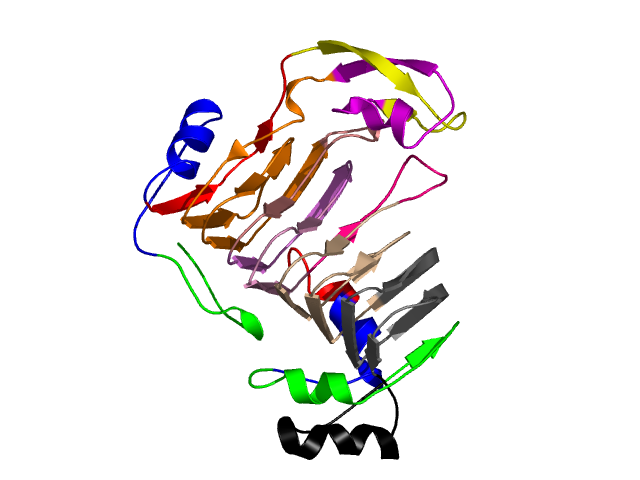
Hierarchical process of splitting
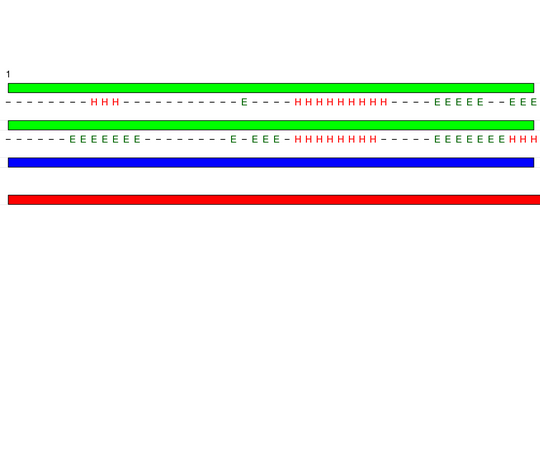
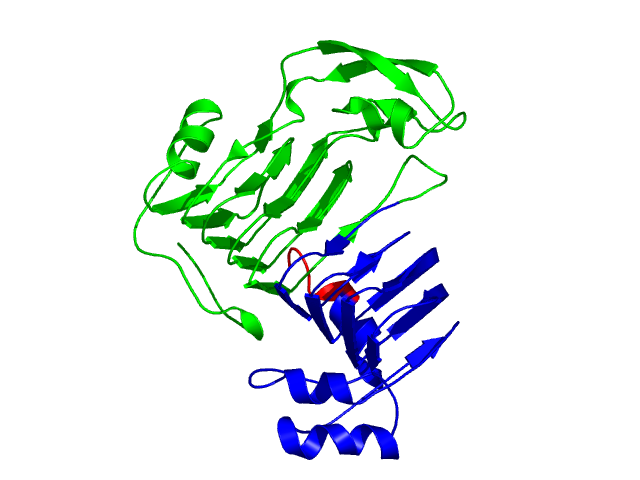
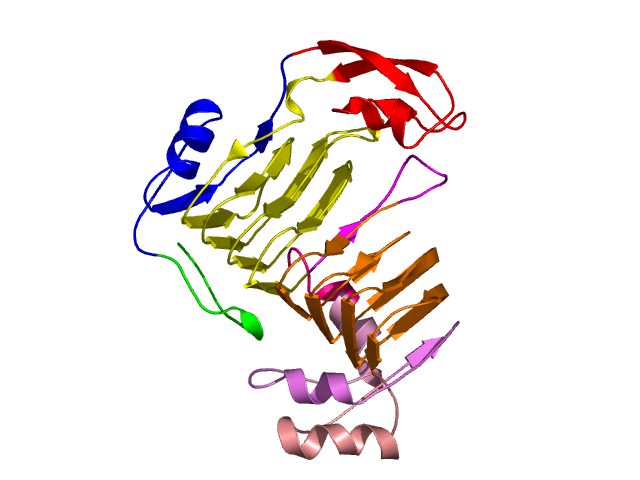
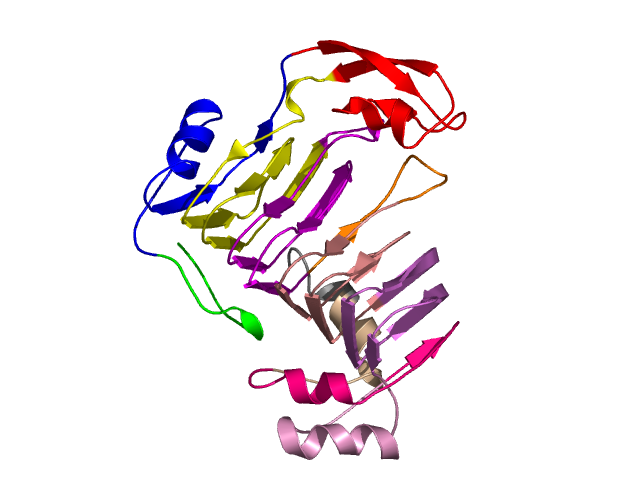
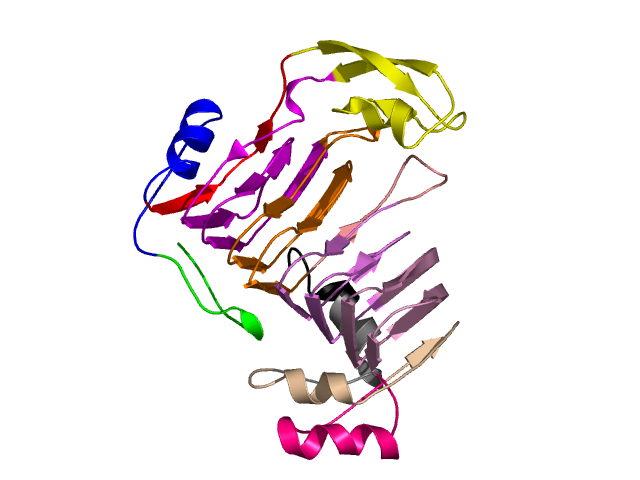
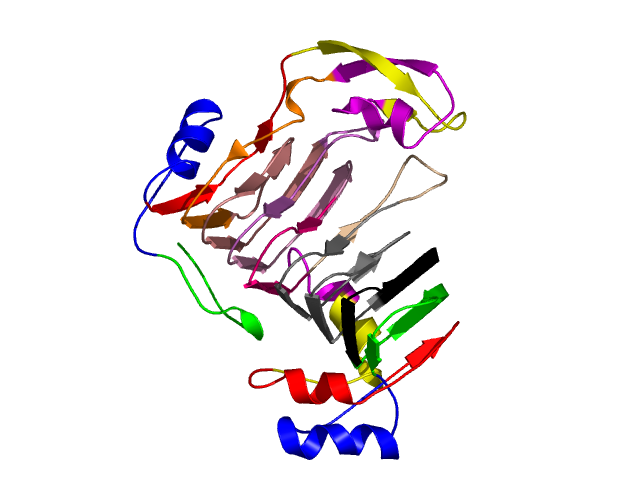
Visualisation of Protein units at all levels
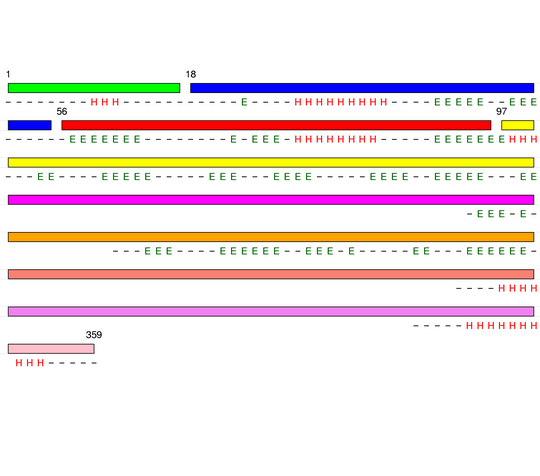
Level 0
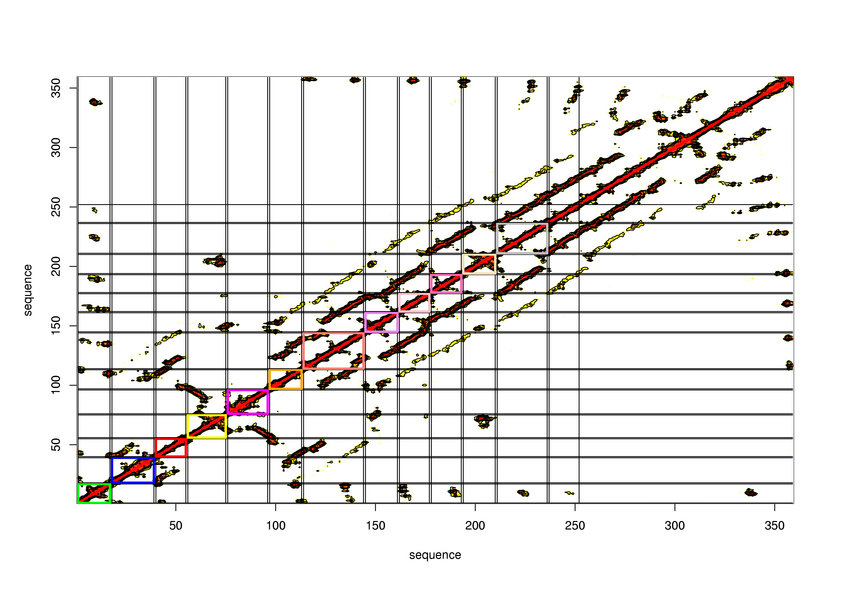
[1-210]:
CI=6.33
[211-351]:
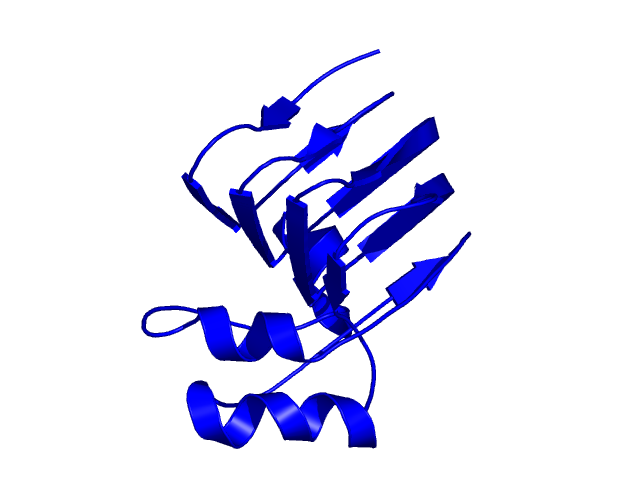
CI=6.21
[352-359
]:
CI=0.22
Level 1
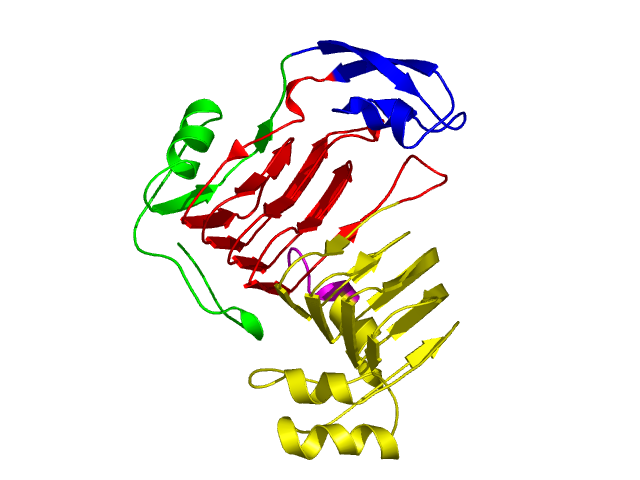
[1-55]:

CI=1.77
[56-96]:
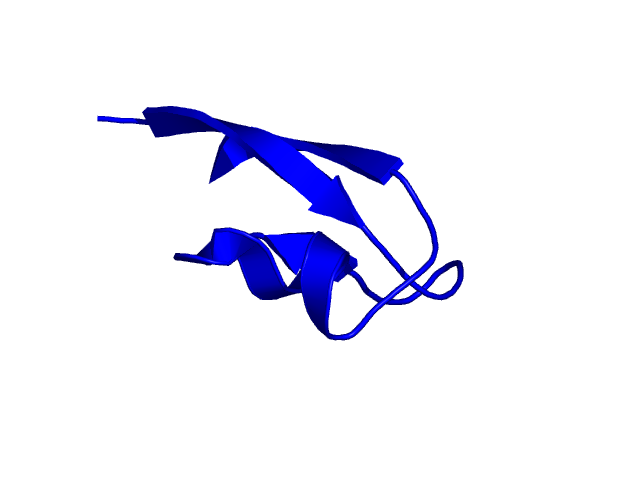
CI=2.72
[97-210]:

CI=6.40
[211-351]:
CI=6.21
[352-359
]:
CI=0.22
Level 2
[1-17]:
CI=0.54
[18-55]:

CI=1.70
[56-96]:

CI=2.72
[97-210]:

CI=6.40
[211-351]:

CI=6.21
[352-359
]:
CI=0.22
Level 3
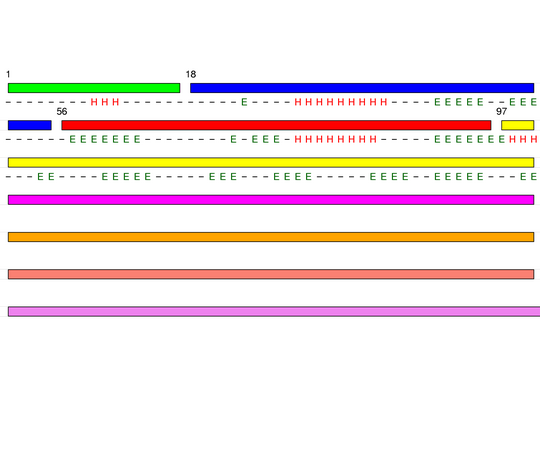
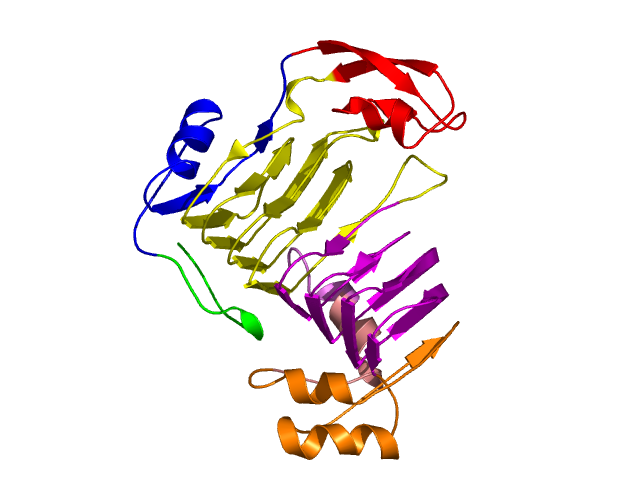
[1-17]:
CI=0.54
[18-55]:

CI=1.70
[56-96]:

CI=2.72
[97-210]:

CI=6.40
[211-292]:

CI=6.63
[293-338]:
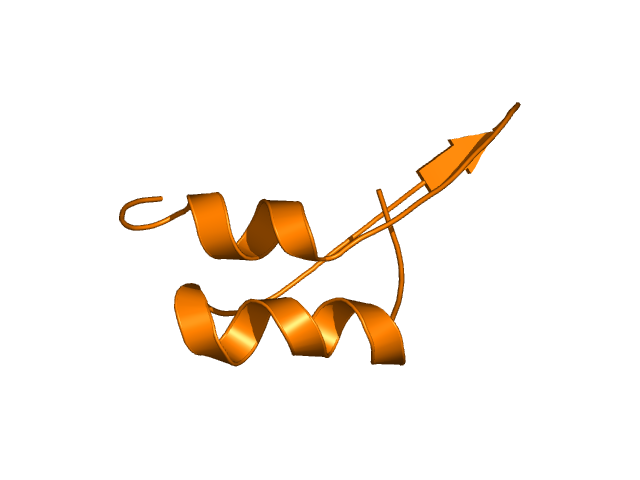
CI=2.16
[339-351]:

CI=0.00
[352-359
]:
CI=0.22
Level 4
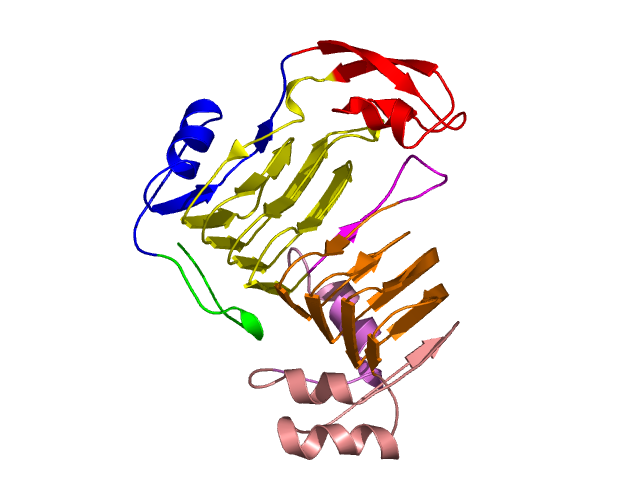
[1-17]:
CI=0.54
[18-55]:

CI=1.70
[56-96]:

CI=2.72
[97-193]:

CI=6.49
[194-210]:

CI=1.54
[211-292]:

CI=6.63
[293-338]:
CI=2.16
[339-351]:
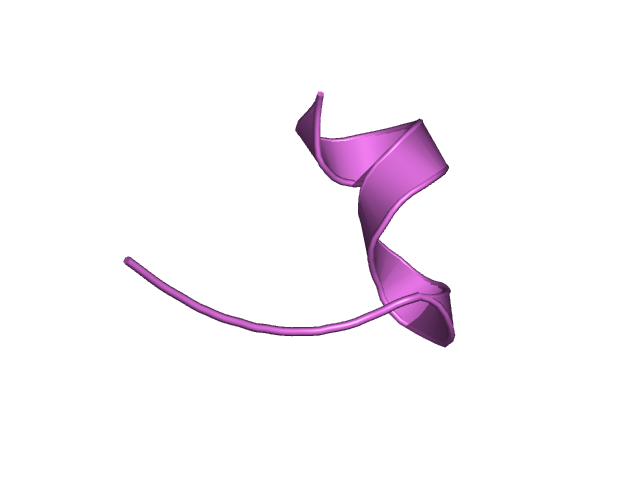
CI=0.00
[352-359
]:
CI=0.22
Level 5
[1-17]:
CI=0.54
[18-55]:

CI=1.70
[56-96]:

CI=2.72
[97-193]:

CI=6.49
[194-210]:

CI=1.54
[211-292]:

CI=6.63
[293-313]:
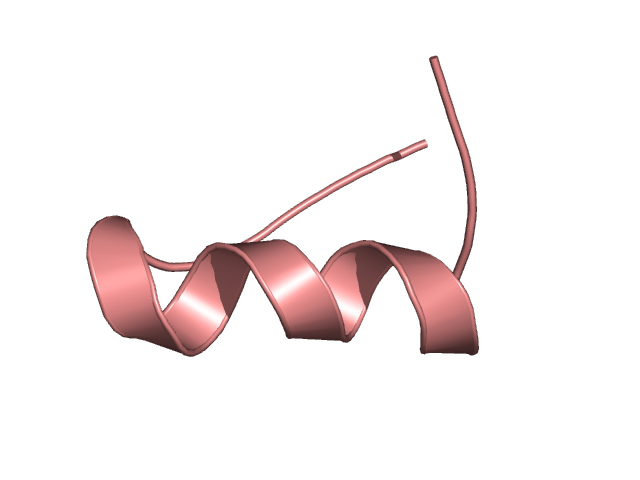
CI=2.05
[314-338]:
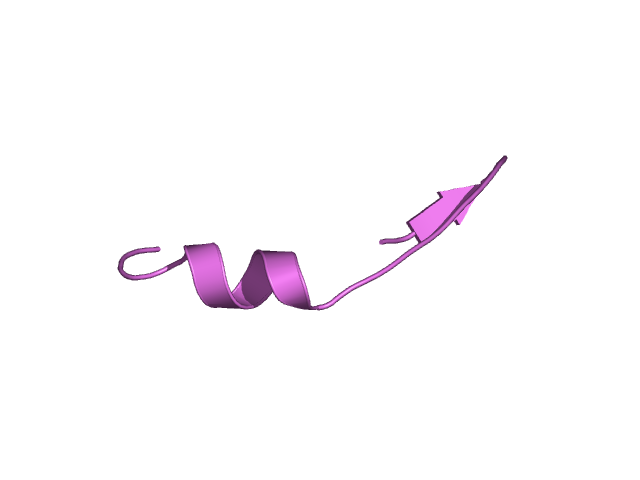
CI=0.07
[339-351]:

CI=0.00
[352-359
]:
CI=0.22
Level 6
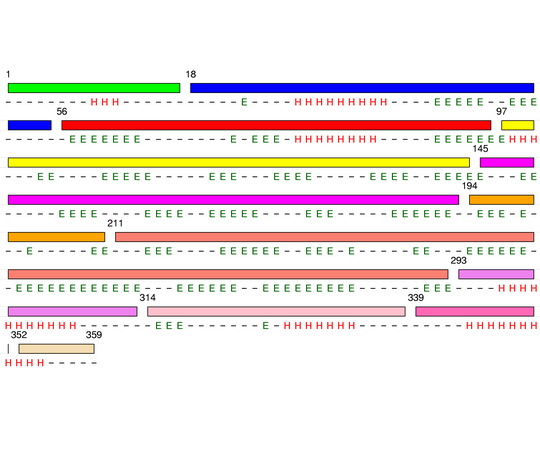
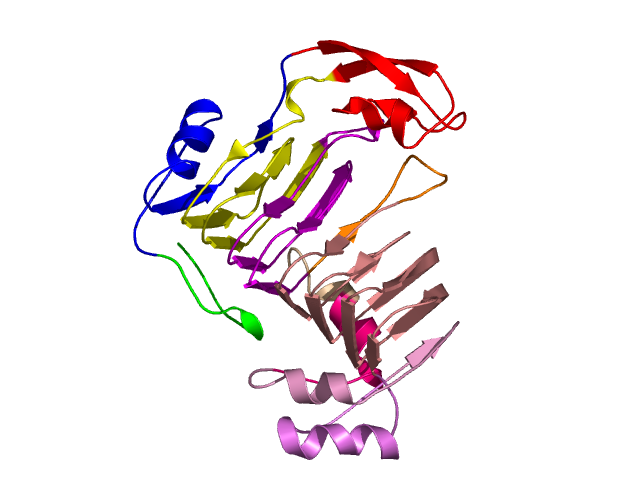
[1-17]:
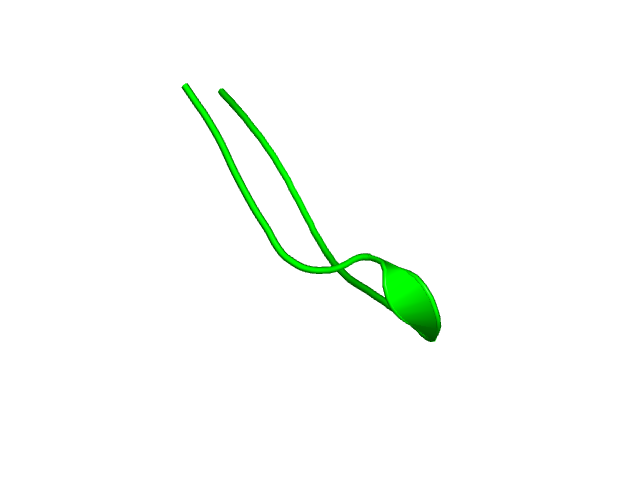
CI=0.54
[18-55]:

CI=1.70
[56-96]:

CI=2.72
[97-144]:
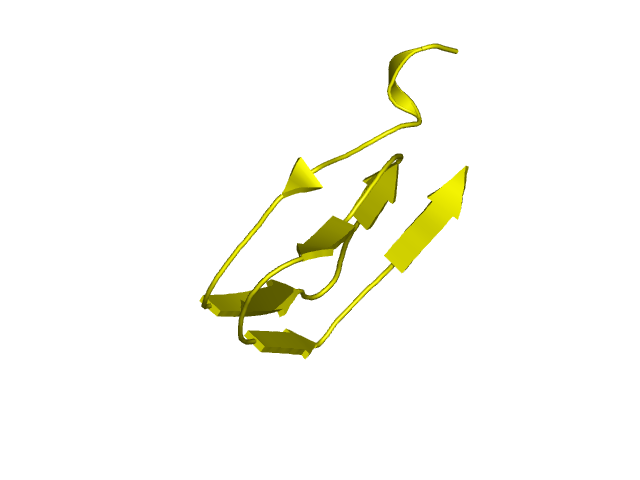
CI=3.64
[145-193]:
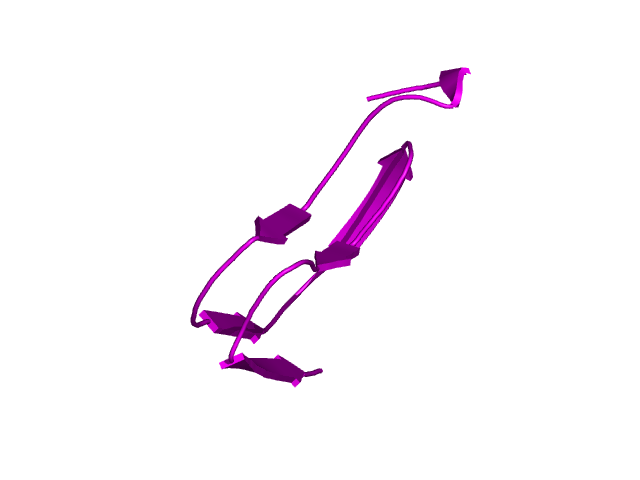
CI=4.24
[194-210]:

CI=1.54
[211-292]:
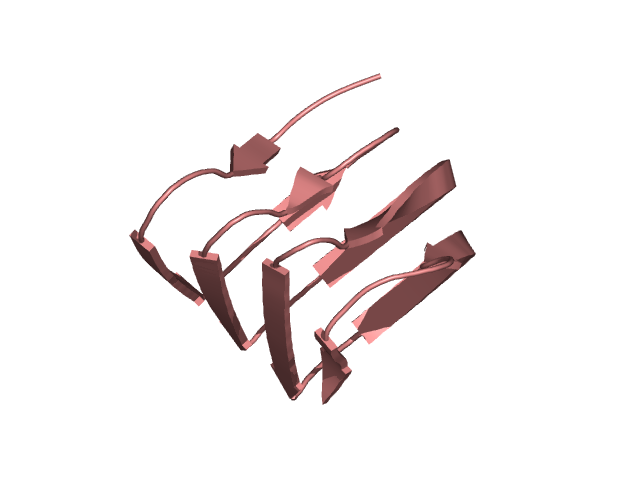
CI=6.63
[293-313]:

CI=2.05
[314-338]:

CI=0.07
[339-351]:
CI=0.00
[352-359
]:
CI=0.22
Level 7
[1-17]:
CI=0.54
[18-55]:

CI=1.70
[56-96]:

CI=2.72
[97-144]:
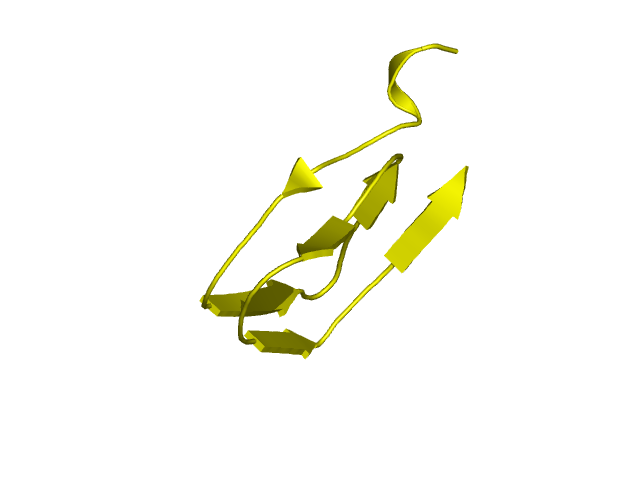
CI=3.64
[145-193]:
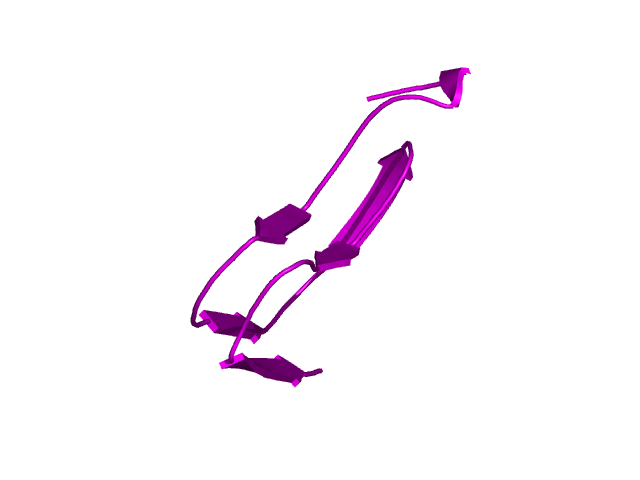
CI=4.24
[194-210]:

CI=1.54
[211-252]:

CI=4.39
[253-292]:

CI=3.46
[293-313]:

CI=2.05
[314-338]:

CI=0.07
[339-351]:
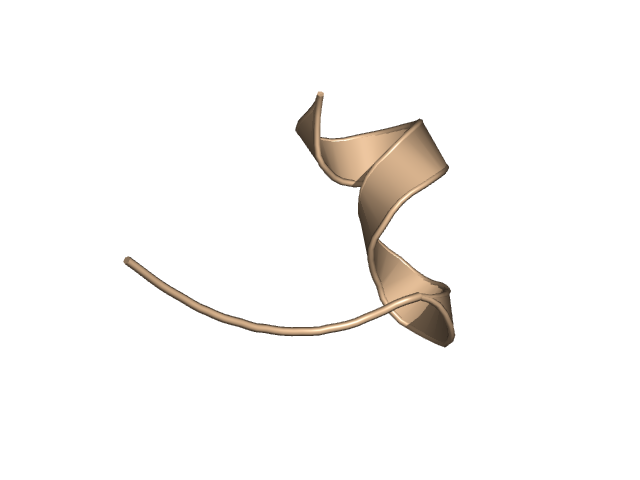
CI=0.00
[352-359
]:
CI=0.22
Level 8
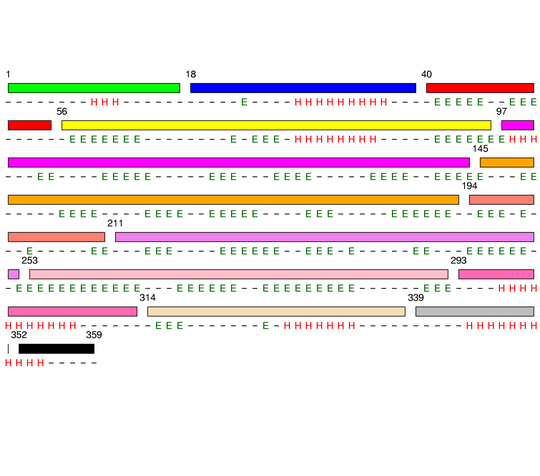
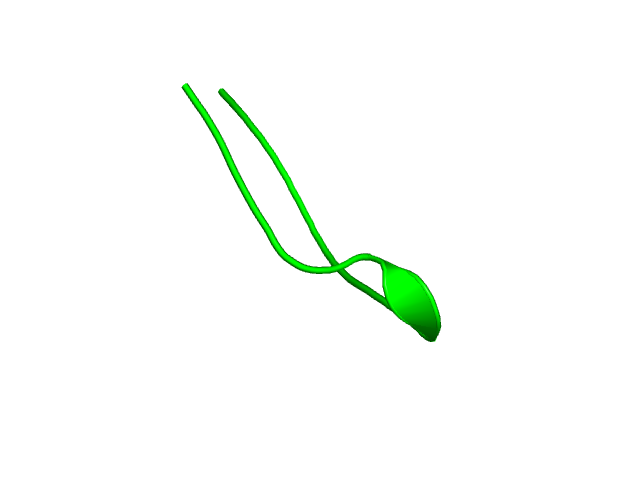
[1-17]:
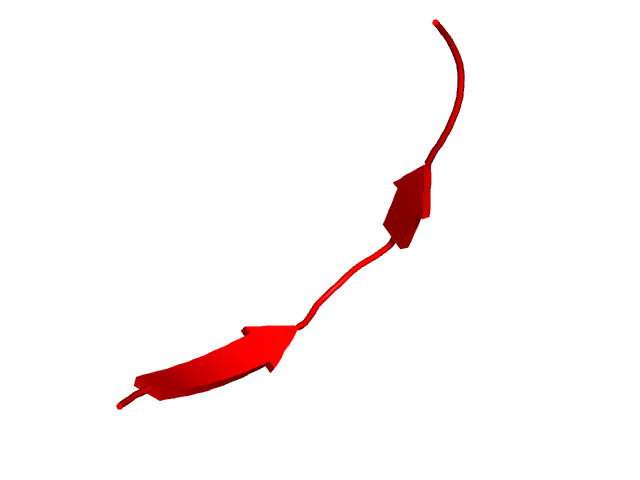
CI=0.54
[18-39]:

CI=0.17
[40-55]:
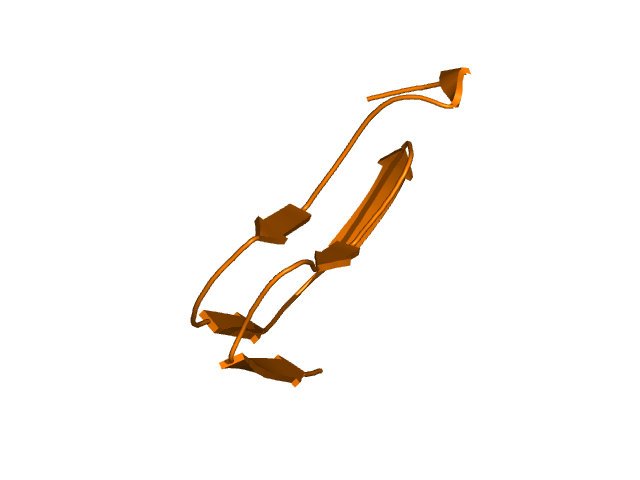
CI=0.00
[56-96]:

CI=2.72
[97-144]:

CI=3.64
[145-193]:
CI=4.24
[194-210]:
CI=1.54
[211-252]:
CI=4.39
[253-292]:

CI=3.46
[293-313]:

CI=2.05
[314-338]:

CI=0.07
[339-351]:

CI=0.00
[352-359
]:
CI=0.22
Level 9
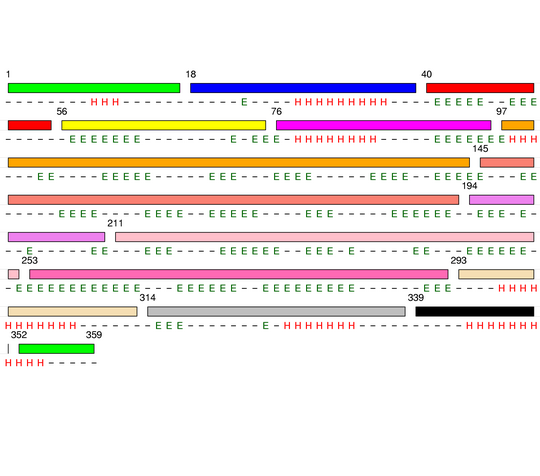
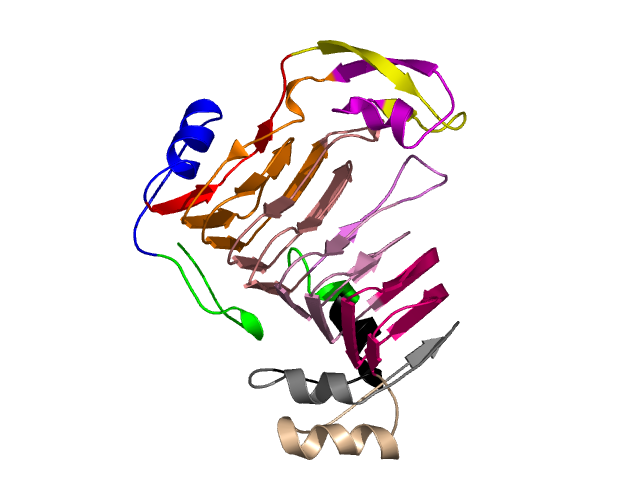
[1-17]:
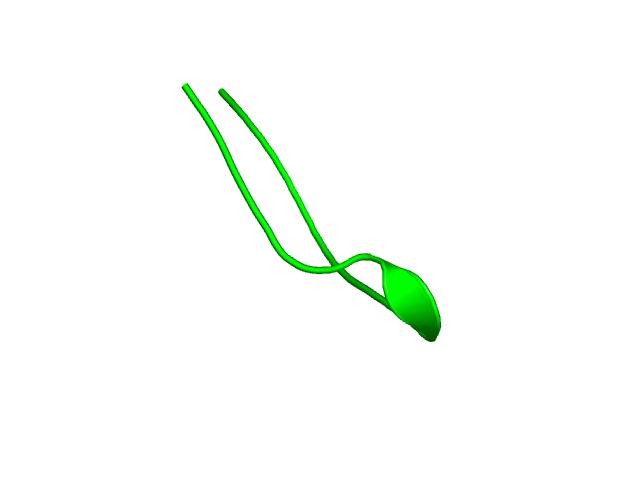
CI=0.54
[18-39]:

CI=0.17
[40-55]:
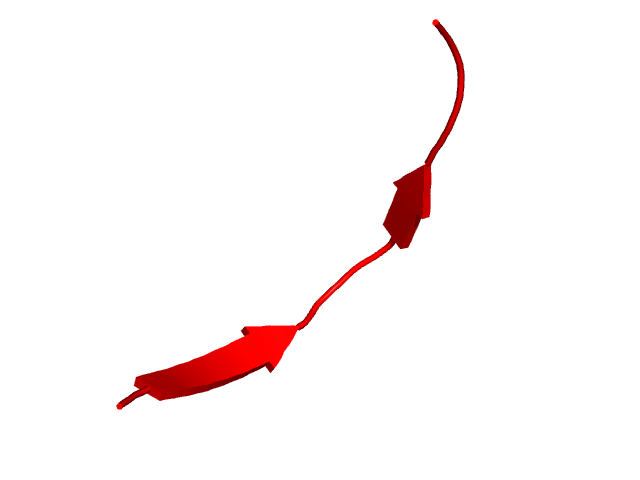
CI=0.00
[56-75]:

CI=1.30
[76-96]:

CI=0.26
[97-144]:

CI=3.64
[145-193]:
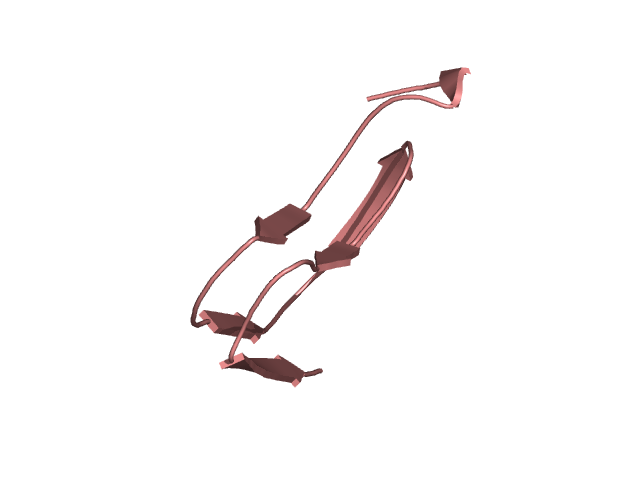
CI=4.24
[194-210]:
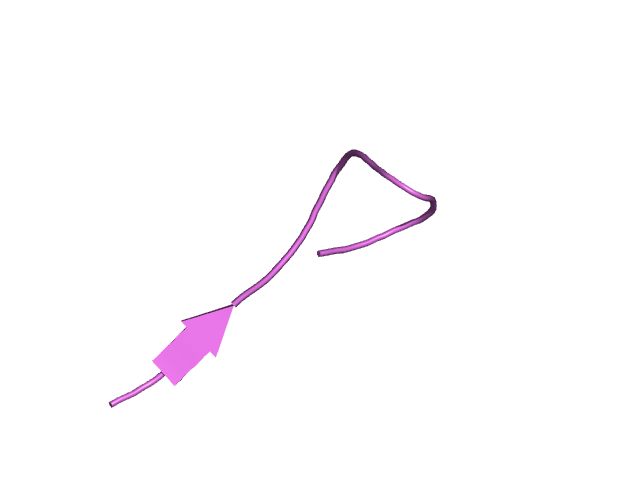
CI=1.54
[211-252]:

CI=4.39
[253-292]:
CI=3.46
[293-313]:

CI=2.05
[314-338]:

CI=0.07
[339-351]:

CI=0.00
[352-359
]:
CI=0.22
Level 10
[1-17]:
CI=0.54
[18-39]:

CI=0.17
[40-55]:
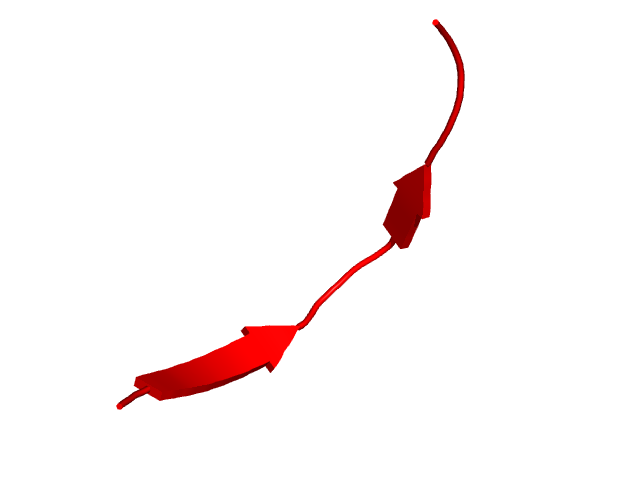
CI=0.00
[56-75]:

CI=1.30
[76-96]:

CI=0.26
[97-144]:

CI=3.64
[145-177]:
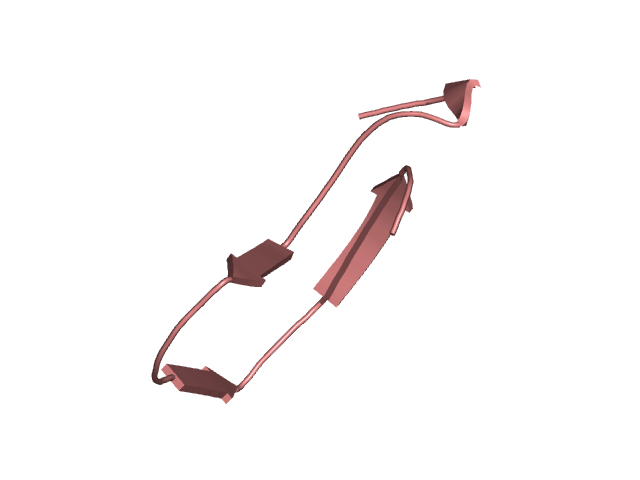
CI=2.47
[178-193]:

CI=0.21
[194-210]:
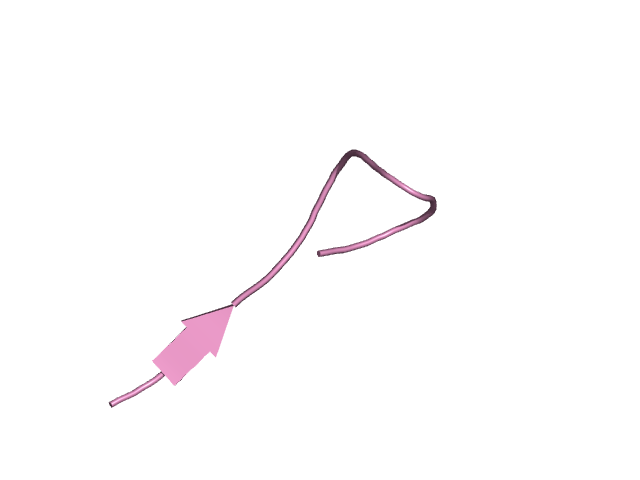
CI=1.54
[211-252]:
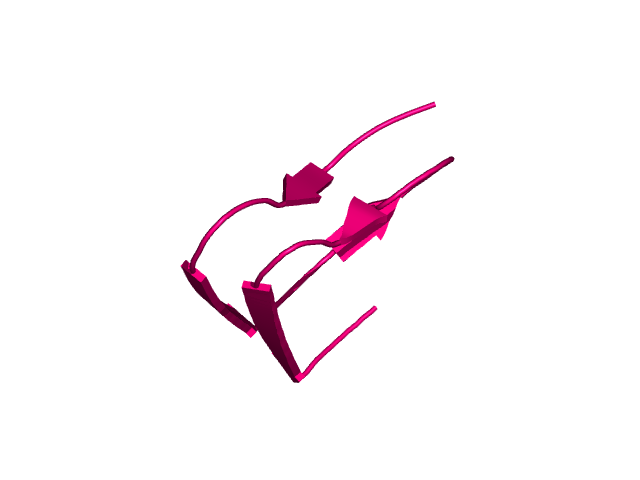
CI=4.39
[253-292]:

CI=3.46
[293-313]:
CI=2.05
[314-338]:

CI=0.07
[339-351]:

CI=0.00
[352-359
]:
CI=0.22
Level 11
[1-17]:
CI=0.54
[18-39]:

CI=0.17
[40-55]:
CI=0.00
[56-75]:

CI=1.30
[76-96]:

CI=0.26
[97-144]:

CI=3.64
[145-161]:

CI=0.65
[162-177]:

CI=0.00
[178-193]:

CI=0.21
[194-210]:
CI=1.54
[211-252]:

CI=4.39
[253-292]:

CI=3.46
[293-313]:
CI=2.05
[314-338]:

CI=0.07
[339-351]:
CI=0.00
[352-359
]:
CI=0.22
Level 12
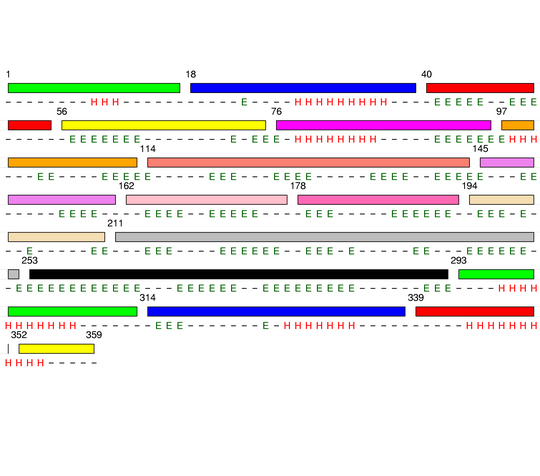
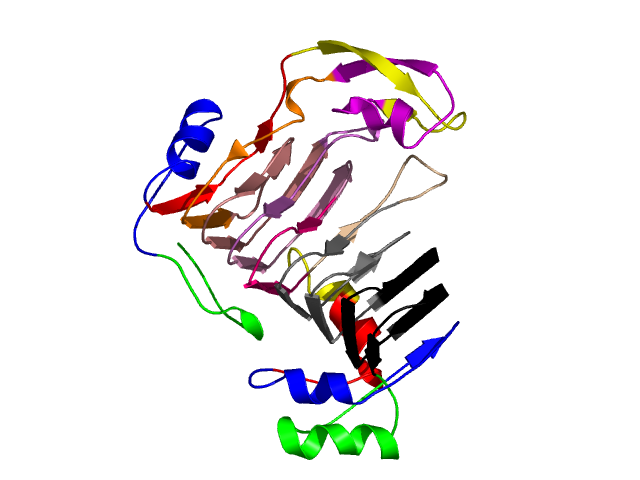
[1-17]:
CI=0.54
[18-39]:

CI=0.17
[40-55]:
CI=0.00
[56-75]:

CI=1.30
[76-96]:

CI=0.26
[97-113]:

CI=0.00
[114-144]:
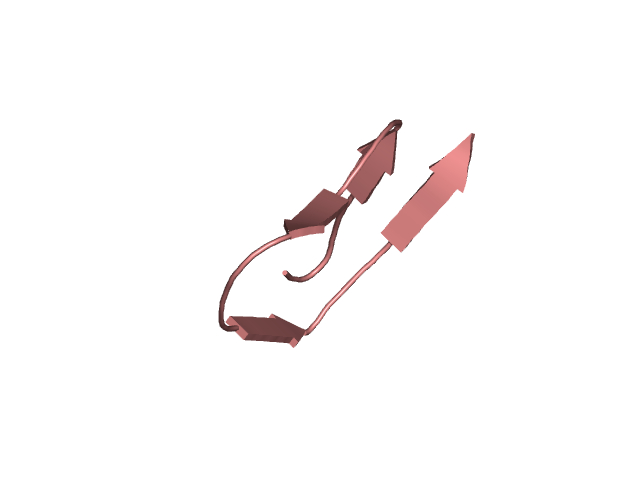
CI=1.95
[145-161]:
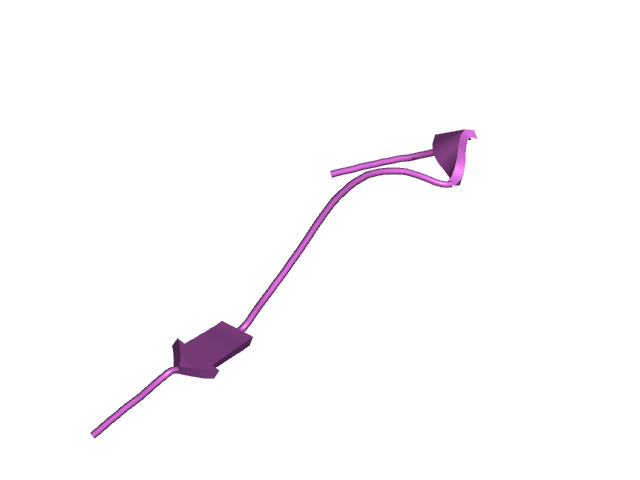
CI=0.65
[162-177]:

CI=0.00
[178-193]:
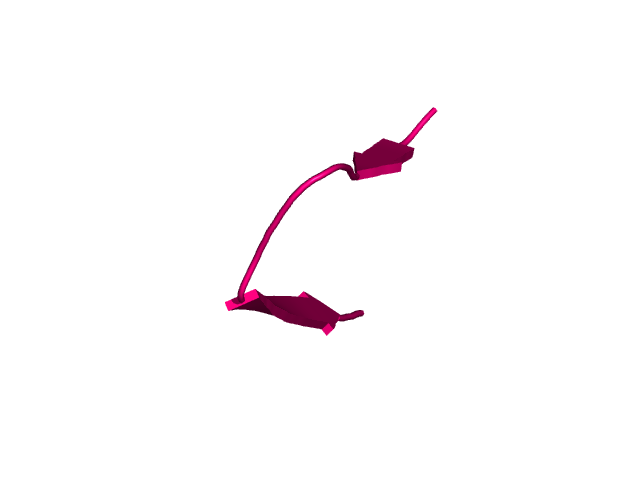
CI=0.21
[194-210]:

CI=1.54
[211-252]:
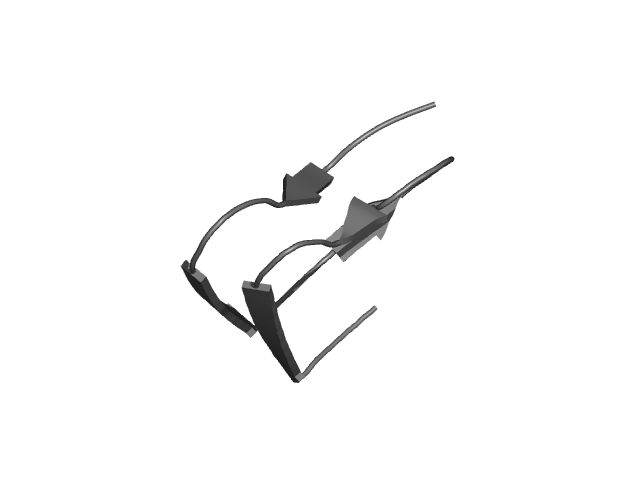
CI=4.39
[253-292]:
CI=3.46
[293-313]:
CI=2.05
[314-338]:

CI=0.07
[339-351]:
CI=0.00
[352-359
]:
CI=0.22
Level 13
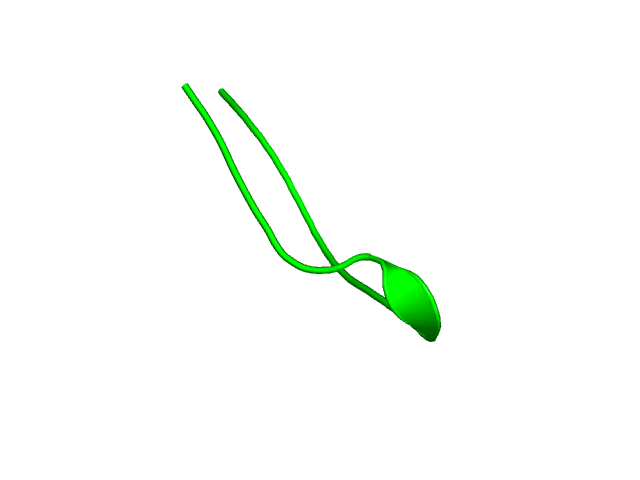
[1-17]:
CI=0.54
[18-39]:

CI=0.17
[40-55]:
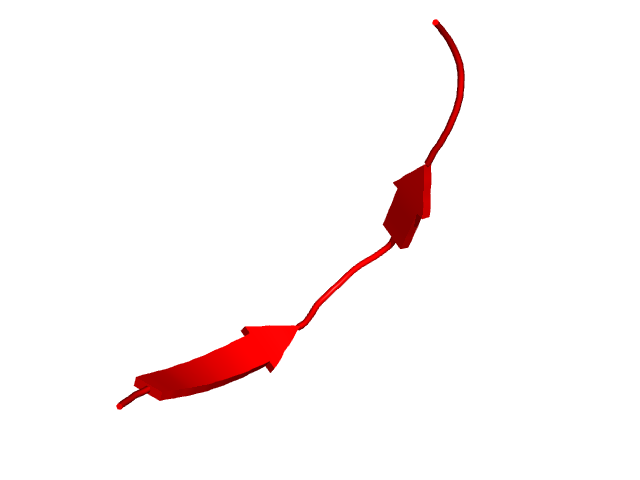
CI=0.00
[56-75]:

CI=1.30
[76-96]:

CI=0.26
[97-113]:

CI=0.00
[114-144]:
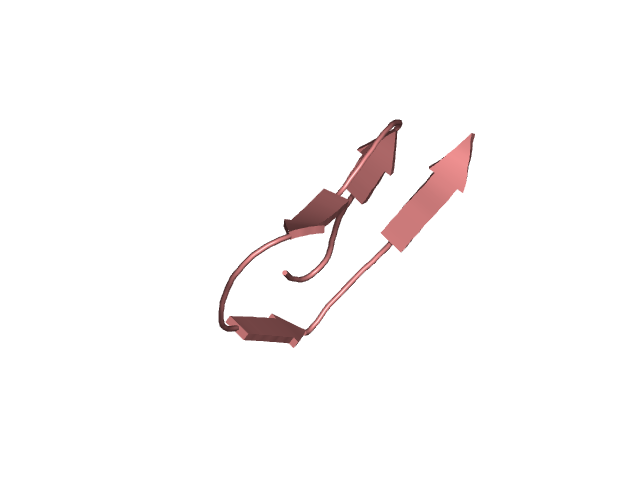
CI=1.95
[145-161]:
CI=0.65
[162-177]:

CI=0.00
[178-193]:
CI=0.21
[194-210]:

CI=1.54
[211-252]:
CI=4.39
[253-271]:
CI=0.19
[272-292]:

CI=0.09
[293-313]:

CI=2.05
[314-338]:

CI=0.07
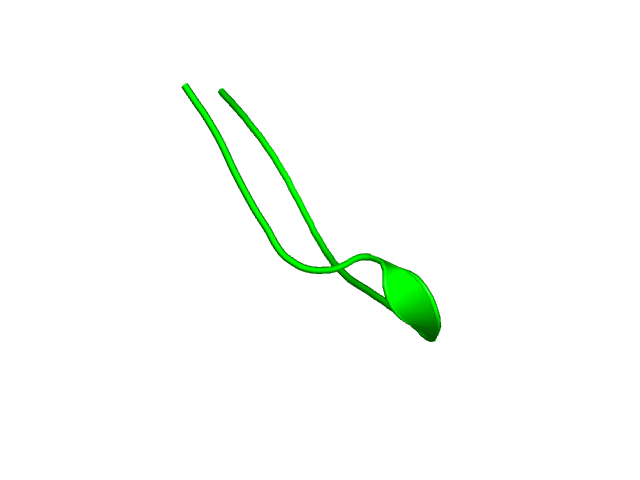
[339-351]:

CI=0.00
[352-359
]:
CI=0.22
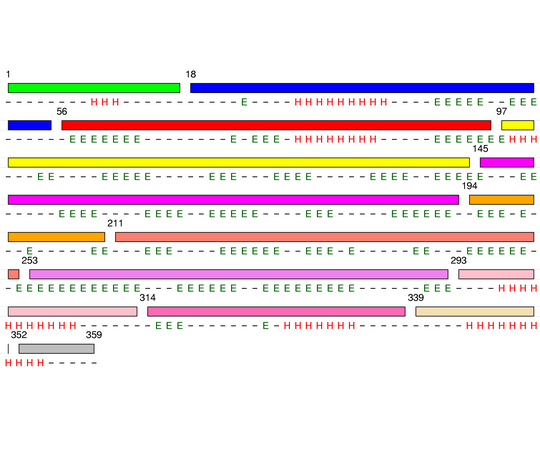
Final level 14
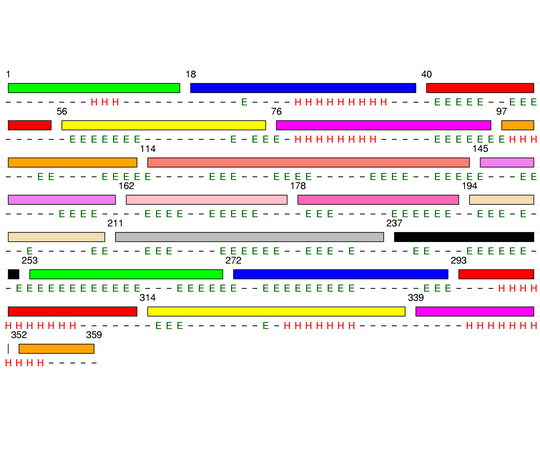
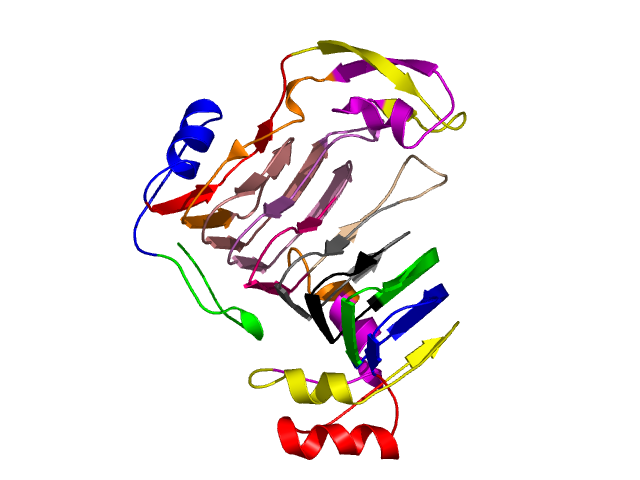
[1-17]:
CI=0.54
[18-39]:

CI=0.17
[40-55]:
CI=0.00
[56-75]:

CI=1.30
[76-96]:

CI=0.26
[97-113]:

CI=0.00
[114-144]:
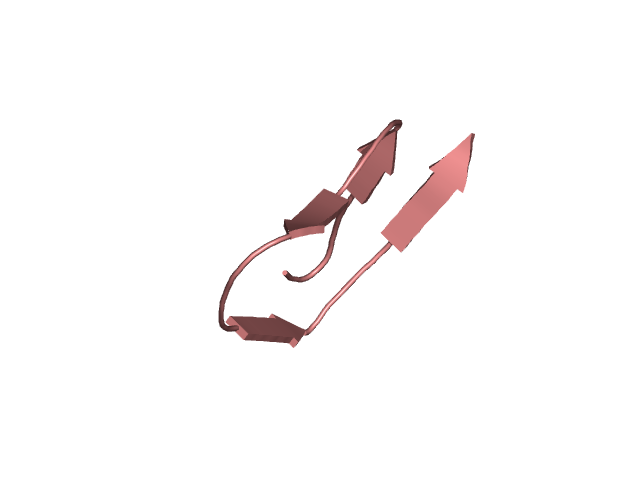
CI=1.95
[145-161]:
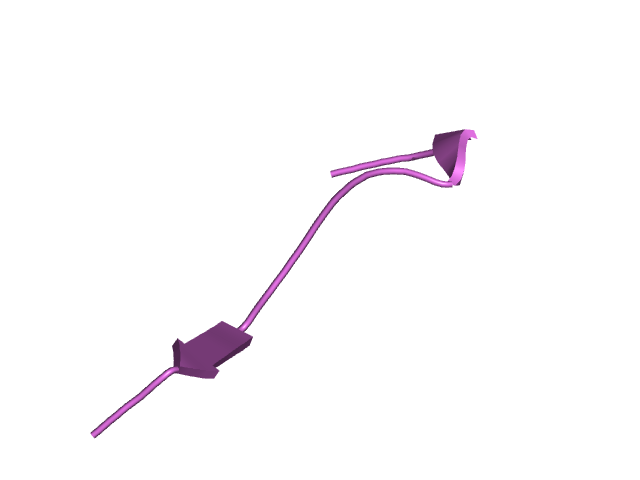
CI=0.65
[162-177]:

CI=0.00
[178-193]:
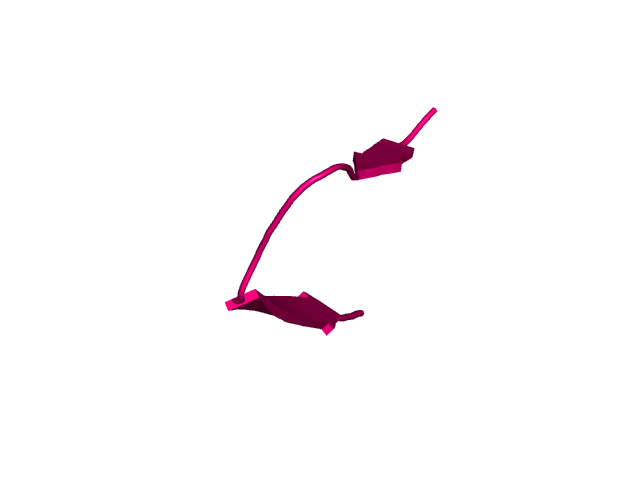
CI=0.21
[194-210]:

CI=1.54
[211-236]:
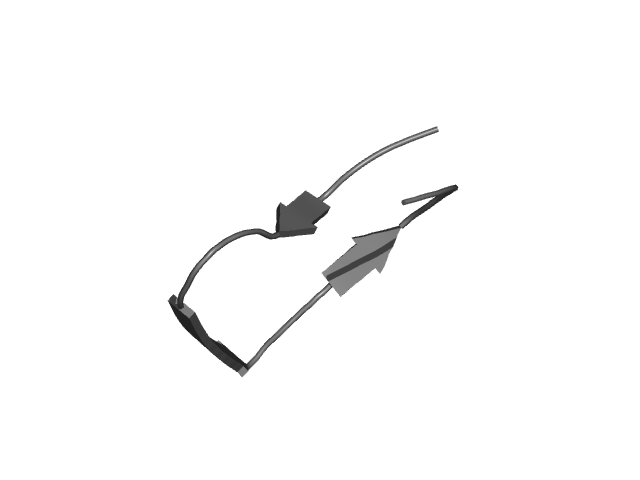
CI=1.09
[237-252]:

CI=0.11
[253-271]:
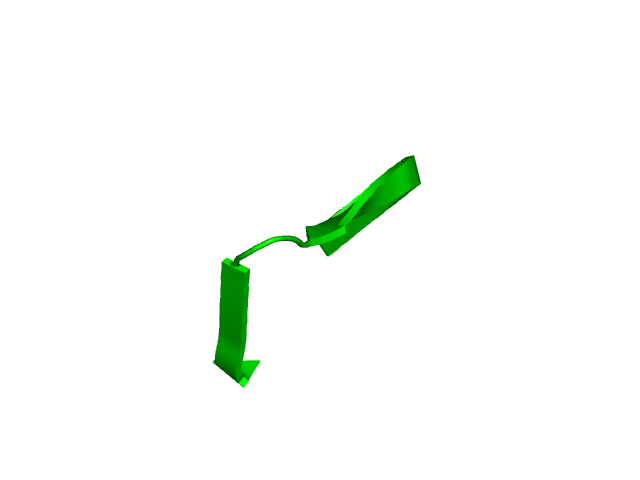
CI=0.19
[272-292]:
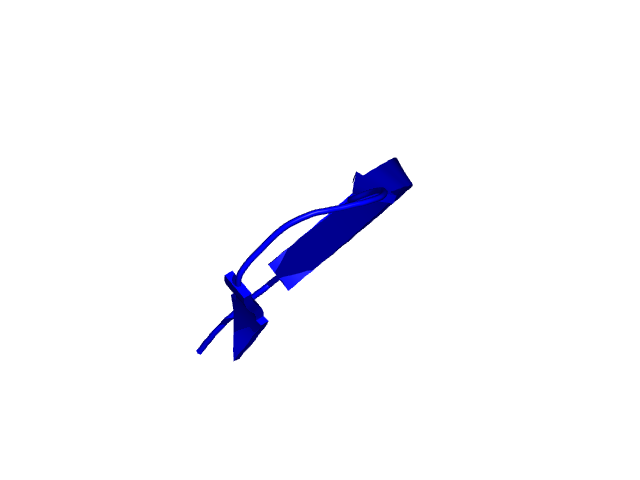
CI=0.09
[293-313]:

CI=2.05
[314-338]:

CI=0.07
[339-351]:
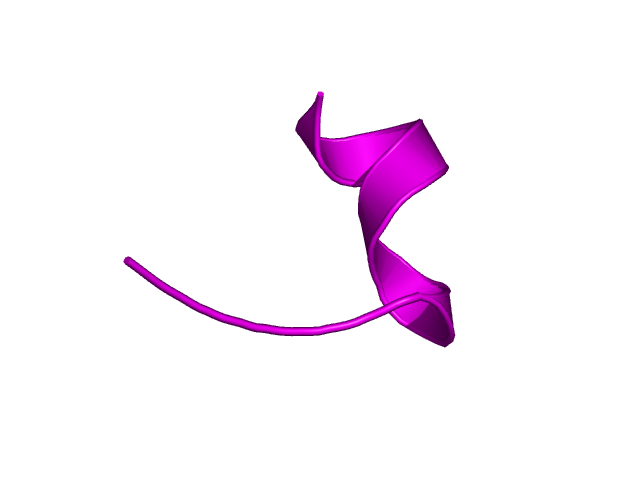
CI=0.00
[352-359
]:
CI=0.22